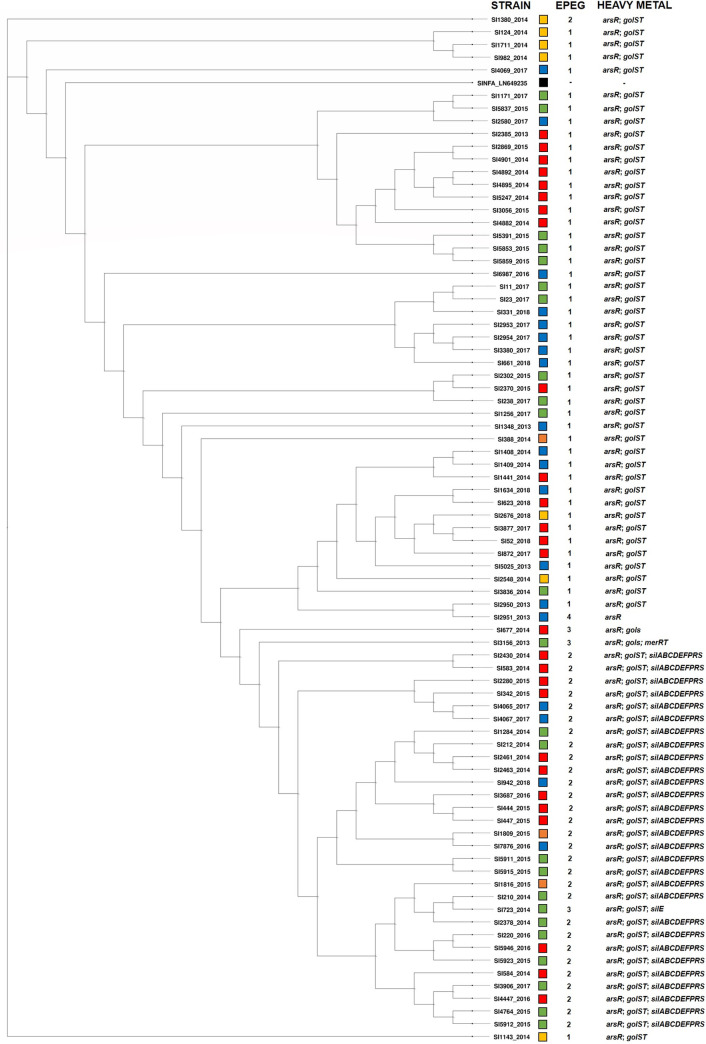
Fig 1. Phylogenetic tree based on the core-genome multi-locus sequence typing (cgMLST) analysis of the 80 whole-genome sequenced S. Infantis strains studied, isolated from food (red squares; n = 27), farm and industry environments (green squares; n = 24), humans (blue squares; n = 19), animals (yellow squares; n = 7) and animal feed (orange squares; n = 3) between 2013 and 2018 in Brazil.
S. Infantis reference strain SINFA LN649235.1 (black square) was included for comparison purposes. Additional information regarding the isolation sources, efflux pump encoding genes (EPEG) and heavy metal tolerance genes are included. Profile 1 (acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdfA, mdtK, msbA, rsmA, sdiA, soxR, soxS); Profile 2 (acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdtK, msbA, rsmA, sdiA, soxR, soxS, tet(A)); Profile 3 (acrA, acrB, baeR, crp, emrB, emrR, golS, hns, kdpE, kpnF, marA, marR, mdtK, msbA, rsmA, sdiA, soxR, soxS); Profile 4 (acrA, acrB, baeR, crp, emrB, emrR, hns, kdpE, kpnF, marA, marR, mdfA, mdtK, msbA, rsmA, sdiA, soxR, soxS).