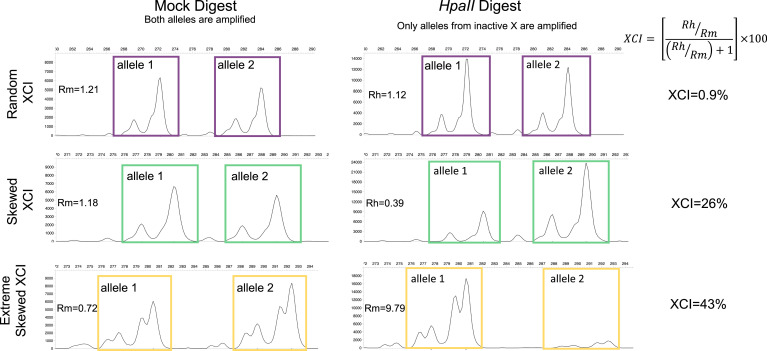
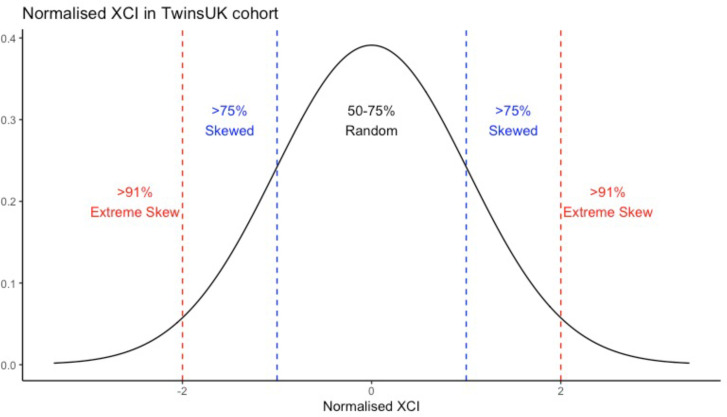
Figure 1. Measuring XCI with the HUMARA assay.
The Human Androgen Receptor Assay (HUMARA) uses methylation-sensitive restriction enzyme digest and PCR to measure skewed X-inactivation. The assay estimates XCI-skew by comparing the relative abundance of allele specific fragments from a mock digest to a methylation-sensitive HpaII digest in which only the alleles from the inactive X are amplified. Representative examples are displayed of fragment analysis of the PCR products for samples with random XCI (top), skewed XCI (middle), and extreme skewed XCI (bottom). The x-axis shows the size, and the y-axis represents the abundance, of the PCR products, respectively. The left panel shows the PCR products after a mock digest with water, resulting in amplification of both alleles regardless of chromosomal inactivation. The right panel shows the PCR products after a restriction enzyme digest with methylation-sensitive enzyme HpaII, resulting in amplification of only the alleles deriving from inactive chromosomes. For each sample, the ratio of the HpaII digested allele products (Rh = allele 1/allele 2) is divided by the ratio of the Mock digest allele products (Rm = allele 1/allele 2) to create a Normalized Ratio (Rn). The XCI percentage is then calculated using the formula [Rn/(Rn +1)] * 100. Images were generated using the Microsatellite Analysis Software on the Thermo Fisher Cloud.