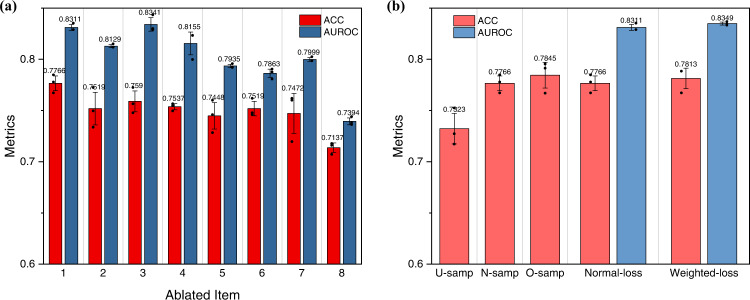
Fig. 4. Validation of DeepPROTACs model and data balance inspection on N = 2832 biologically independent samples over 3 independent experiments.
Data are presented as mean values ± SD. The statistical test used for data analysis is paired t test with two-sided and no adjustments were made for multiple comparisons. Source data are provided as a Source Data file. a Ablation experiments on DeepPROTACs model. Ablated item: 1 - none, 2 - ligase pocket, 3 - E3 ligand, 4 - POI pocket, 5 - warhead, 6 - linker, 7 - ligase pocket and E3 ligand, 8 - POI pocket and warhead. b Data balance experiments on DeepPROTACs model. Under-sampling (U-samp): deleting redundant inactive samples (the number of active and inactive samples is 988:988). Normal-sampling (N-samp): adopting original samples (the number of active and inactive samples is 988:1844). Over-sampling (O-samp): repeated sampling part of active samples (the number of active and inactive samples is 1844:1844). Weighted-loss: the weight of loss corresponding to the active samples was multiplied by 2, while the weight of loss corresponding to the inactive samples was unchanged.