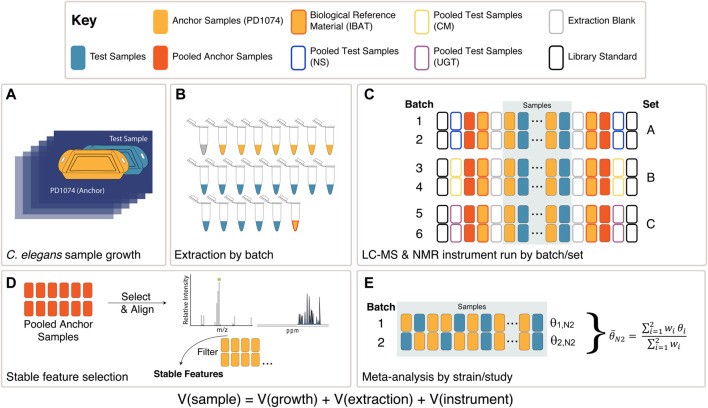
FIGURE 2.
Experimental design overview. (A) Each C. elegans LSCP was grown and harvested with at least one PD1074 sample (sample growth variation captured) (Supplementary Figure S1). (B) PD1074 samples and test strains (NS, CM mutants, or UGT mutants), IBAT references, and extraction blanks were included in each batch for LC-MS or NMR (batch preparation variation captured). (C) A total of six batches in three sets were collected. Instrument controls, library standards, and pooled PD1074 samples were included at the start and end of each run (instrumentation variation captured) were in each run. Independent samples from each test strain were collected in two independent sequential batches. (D) In LC-MS, PD1074 spectral features were identified from PD1074 pools and retained if they were present above the level of the extraction blank in 100% of the individual PD1074 spectra. In NMR, semi-automated peak-picking and binning were performed to extract peak heights and identify stable peaks present in PD1074 samples. (E) Meta-analysis models to identify differences between test strains and PD1074.