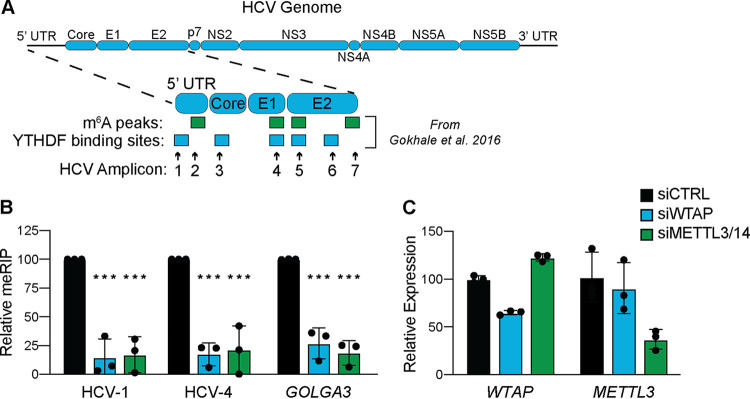
FIG 2.
WTAP and METTL3 + 14 are essential for m6A modification of HCV RNA. (A) Illustration of the HCV RNA genome with amplicons measured in this study and m6A peaks or YTHDF protein binding sites identified in reference 5. UTR, untranslated region. (B and C) Relative meRIP enrichment of the indicated viral or cellular amplicons from Huh7 cells treated with the indicated siRNAs and infected with HCV (48 h, MOI of 0. 3) (B), as determined as the percentage of input under each condition normalized to an m6A RNA spike-in with siCTRL samples normalized to 100%, and RT-qPCR analysis of indicated genes (C), relative to 18S rRNA. For panel B, graph shows mean ± SD (n = 3 biological replicates), while panel C is representative. Data were analyzed by two-way ANOVA with Šidák’s multiple-comparison test (***, P < 0.001).