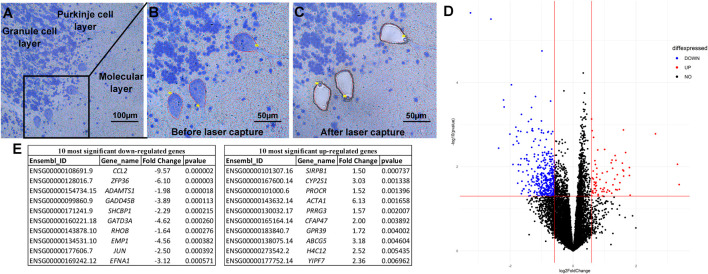
FIGURE 1.
Laser capture microdissection and differential expression analysis. Tissue is stained with a rapid Nissl protocol, which clearly delineates Purkinje cells based on the consistent architecture of the cerebellar cortex and large cell bodies (A). The Zeiss RoboPalm software is used to manually outline individual thinly sliced (8 µm) Purkinje cells (B) and specifically removes the cell body, leaving the other cells and tissue behind (C). After RNA extraction and sequencing, Principle Linear regression models within DESeq2 were used to test differentially expressed genes between individuals with ASD and controls, correcting for covariates of age, self-reported ethnicity, sex, and post-mortem interval (D). The top ten significant up- and down-regulated genes are provided (E).