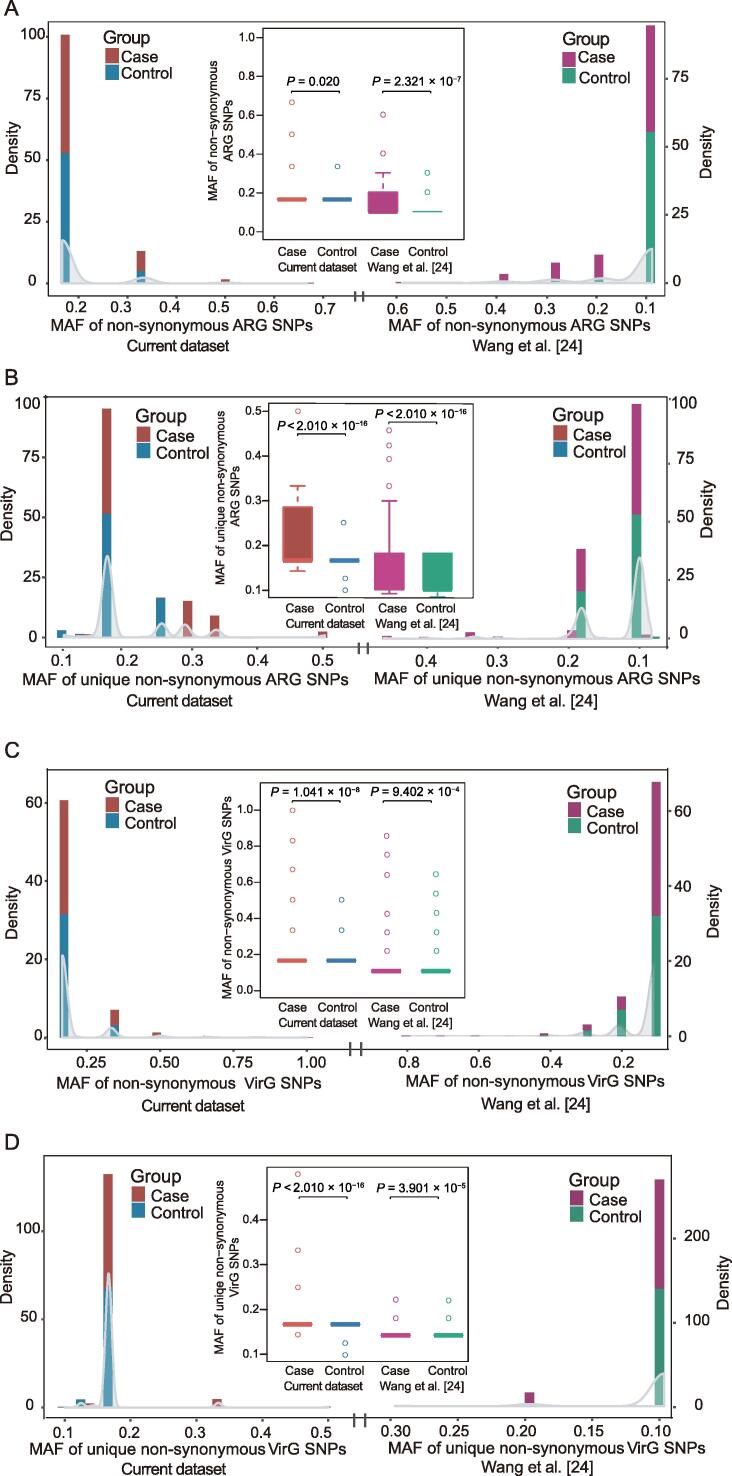
Figure 4.
Comparison of non-synonymous SNPs in ARGs and VirGs between periodontitis and control samples from both datasets
A. Distribution of the MAF of non-synonymous SNPs in ARGs from both the current study and a study by Wang and colleagues [24]. Comparison of the MAF of non-synonymous SNPs in ARGs between periodontitis and control samples in both datasets. B. Distribution of the MAF of unique non-synonymous SNPs in ARGs from both the current study and a study by Wang and colleagues [24]. Comparison of the MAF of unique non-synonymous SNPs in ARGs between periodontitis and control samples in both datasets. C. Distribution of the MAF of non-synonymous SNPs in VirGs from both the current study and a study by Wang and colleagues [24]. Comparison of the MAF of non-synonymous SNPs in VirGs between periodontitis and control samples in both datasets. D. Distribution of the MAF of unique non-synonymous SNPs in VirGs from both the current study and a study by Wang and colleagues [24]. Comparison of the MAF of unique non-synonymous SNPs in VirGs between periodontitis and control samples in both datasets. All P values were calculated using Wilcoxon rank sum test. SNP, single-nucleotide polymorphism; ARG, antibiotic resistance gene; VirG, virulence gene; MAF, minor allele frequency.