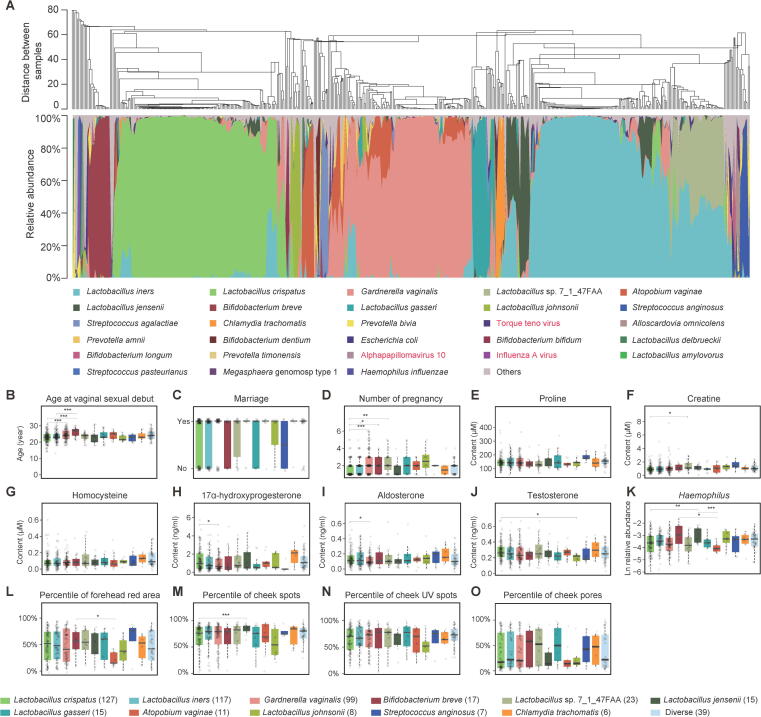
Figure 2.
Vagino-cervical microbiome of the initial study cohort
A. The microbial composition in each sample at the species level according to MetaPhlAn2 is shown. The dendrogram in (top) was a result of a centroid linage hierarchical clustering based on Euclidean distances between the microbial composition proportion. Black and red taxa labels denote bacteria and viruses, respectively. B.–O. Specific multi-omic factors showing significant differences among 12 key vagino-cervical microbiota types estimated by GLM likelihood ratio test (P < 0.05, 554 comparisons). Colors of the bars in (B–O) denote the microbiota types as listed at the bottom of the figure. The number of samples per microbiota type is labeled in the parentheses. The horizontal lines with P values mark the pairwise post hoc comparisons using Student’s t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). The genus in (K) belongs to fecal microbiome. Boxes denote the IQR between the first and third quartiles (25th and 75th percentiles, respectively), and the line inside the boxes denotes the median. The whiskers denote the lowest and highest values within 1.5 times the IQR from the first and third quartiles, respectively. GLM, generalized linear model; IQR, interquartile range.