Abstract
COVID-19 had caused the whole world to come to a standstill. The current detection methods are time consuming as well as costly. Using Chest X-rays (CXRs) is a solution to this problem, however, manual examination of CXRs is a cumbersome and difficult process needing specialization in the domain. Most of existing methods used for this application involve the usage of pretrained models such as VGG19, ResNet, DenseNet, Xception, and EfficeintNet which were trained on RGB image datasets. X-rays are fundamentally single channel images, hence using RGB trained model is not appropriate since it increases the operations by involving three channels instead of one. A way of using pretrained model for grayscale images is by replicating the one channel image data to three channel which introduces redundancy and another way is by altering the input layer of pretrained model to take in one channel image data, which comprises the weights in the forward layers that were trained on three channel images which weakens the use of pre-trained weights in a transfer learning approach. A novel approach for identification of COVID-19 using CXRs, Contrast Limited Adaptive Histogram Equalization (CLAHE) along with Homomorphic Transformation Filter which is used to process the pixel data in images and extract features from the CXRs is suggested in this paper. These processed images are then provided as input to a VGG inspired deep Convolutional Neural Network (CNN) model which takes one channel image data as input (grayscale images) to categorize CXRs into three class labels, namely, No-Findings, COVID-19, and Pneumonia. Evaluation of the suggested model is done with the help of two publicly available datasets; one to obtain COVID-19 and No-Finding images and the other to obtain Pneumonia CXRs. The dataset comprises 6750 images in total; 2250 images for each class. Results obtained show that the model has achieved 96.56% for multi-class classification and 98.06% accuracy for binary classification using 5-fold stratified cross validation (CV) method. This result is competitive and up to the mark when compared with the performance shown by existing approaches for COVID-19 classification.
Keywords: COVID-19, Homomorphic Transformation Filter, Deep CNN, VGG, ANOVA
1. Introduction
The first case of Severe Acute Respiratory Syndrome Coronavirus 2, abbreviated as SARS-CoV-2, was identified and reported in December 2019, in the city of Wuhan, China [1]. The World Health Organization (WHO) named this infection caused by SARS-CoV-2 as coronavirus disease 2019, abbreviated as COVID-19. COVID-19 is a highly contagious disease with various symptoms ranging from cough, fever, fatigue, etc. The general population is highly vulnerable to infection caused by this virus. Since the pandemic's outbreak and rapid spread, it has become clear that disease prognosis is heavily influenced by multi-organ involvement [2]. Death was caused by acute respiratory distress syndrome, heart failure [3], renal failure [4], liver damage [5], hyper-inflammatory shock [6], and multi-organ failure [7]. Due to the limited number of testing facilities available and the disease's early stages' low prevalence of positive symptoms, the currently available RT-PCR method used for detection and identification of COVID-19, which stands for Reverse Transcription Polymerase Chain Reaction, poses some drawbacks hence creating the need for other alternatives and options. Some other methods of detection include Computer Tomography (CT) scans and Chest X-rays (CXRs). These are important since a confirmed COVID-19 patient may or may not have a normal chest scan during the initial stages of contracting the infection [8].
CXR is a common, more affordable alternative to CT scans. It also takes lesser time for generation which serves as an added advantage to its utility. Recent technologies, particularly artificial intelligence (AI) tools, have been investigated for tracking the transmission of the coronavirus, identifying individuals at high risk of mortality, and diagnosing patients with the condition (see Table 1 ).
Table 1.
Hyperparameter table of the suggested GrayVIC model.
| Hyperparameters | Values |
|---|---|
| COVID-19 instances | 2250 |
| Pneumonia instances | 2250 |
| No-findings instances | 2250 |
| Image resolution | 64 × 64 × 1 |
| Learning rate | 10-3 |
| Minimum LR | 10-6 |
| Batch size | 64 |
| Epochs | 100 |
| Optimizer | Adaptive Moment Estimation (Adam) |
| Loss function | Categorical cross-entropy |
Artificial Intelligence has huge underlying potential in curbing the COVID-19 pandemic with the help of successful practical implementations using CXRs and CT scans [9]. The usage of pre-trained architectures like Deep Convolutional Neural Network (DCNN), viz. GoogleNet, NASNet, VGGNet, and DenseNet are used for the implementation of this application. In addition, the model achieves higher accuracy as image processing techniques improve.
Sometimes, a trained expert in this field might miss some attributes which confirm the infection, either due to higher traffic of patients or fatigue, and might need quick detection and identification. This is where a deep learning model can be employed for better and faster interpretation for detection of the infection. Almost all the related works have used a pre-trained CNN model that was trained on three channel (RGB) images to obtain the weights which is not appropriate when it comes to X-ray images as they are single channel (grayscale) images. This paper’s goal is to propose a novel architectural model inspired from VGG16 architecture to classify X-ray (grayscale) images according to the disease classes based on both binary and multi-class classification. In order to train the model more efficiently, pre-processing combination techniques have been employed, specifically, CLAHE and Homomorphic Transformation. The classifier model is built from scratch hence it is entirely trained on the image dataset. To avoid the erratic fluctuation noticed in the validation accuracy during the training phase, ReduceLRonPlateau method has been used. The authors have ensured the robustness of model on the public dataset by following the stratified 5-fold cross-validation methodology. All necessary performance metrics have been estimated for the comparison purposes and tabulated properly in the results section.
The motivations and contributions of the proposed work are discussed in Section 2. In Section 3, we discuss the existing methods and related works employed for identifying the infection caused by the SARS-CoV-2 virus using Chest X-rays. The dataset used for the proposed work is elaborated upon in Section 4. Section 5 describes the approach used by us for COVID-19 detection using a deep learning Convolutional Neural Network model in detail. Section 6 and 7 explain and discuss the results obtained and conclude the paper respectively.
2. Motivation and contributions
The developed deep CNN model is inspired by VGG architecture. Customized CNN models with the help of transfer learning architectures shows promising results when comes to efficiency [10]. The customized CNN models have seen to have better time complexity, faster learning rate [11]. The model employs no pre-trained models’ weights and provides promising output with very little training time compared to other existing models. The proposed model takes CXRs as input, making it cost effective since CT scans are unreasonable and may not be accessible at an individual level. Then a random selection of these images is provided to the model which works on a single channel. The testing is done in two ways; first, which comprises binary classification where only two modes, COVID-19 and No-Findings, are taken into consideration. The second method involves the classification of CXRs across three classes, namely, No-Findings, COVID-19, and Pneumonia. The image processing techniques used, viz. CLAHE and Homomorphic Transformation filter help in improvising its contrast and plummeting its dynamic range. These image processing techniques incorporated into the proposed model produce a more robust diagnosis for COVID-19. The novel aspect of this work is the application of Homomorphic Transformation Filter as an image processing approach on CXRs as well as the designing a VGG inspired Deep CNN model from scratch that requires less training time than models using the same range of datasets. The significant contributions made by this paper include the following:
-
1.
CLAHE + Homomorphic Transformation Filter as image processing technique on CXRs.
-
2.
A novel VGG inspired deep CNN model consisting of 22 layers and the inputs fed to the model have the shape of 64 × 64 × 1.
-
3.
Different hyper parameter tuning methodologies are used to examine the potential of the proposed model for the task of multi as well as binary-class classification of CXRs.
-
4.
Assessment of model’s robustness with the help of 6750 images.
3. Related works
Currently, Deep Learning is adopted in the field of medical imaging. This includes analysis of medical images, radiomics, etc. Deep Learning is mainly used because of its prominent and reliable results [12]. Since there is no restriction to the kind of data that can be used, deep learning is appropriate to cater to diverse information and data, in order to make predictions [13]. Due to the prolonged pandemic caused by COVID-19, it has become a necessity to come up with a technology that uses deep learning concepts and techniques in order to detect COVID-19 faster and with a higher degree of accuracy since current testing methods are expensive and time consuming. CXRs and CT scans can be helpful in achieving this. Since CXRs are more affordable as compared to CT scans, they are a better alternative.
Aslan, M. F. et al. worked on binary classification techniques and observed the DenseNet-SVM structure [14] to be the best one with an accuracy of about 96.29 %. They achieved an average accuracy of 95.21 % by using a total of eight different SVM based CNN models. Alakus, T. B. et al. validated their LSTM deep learning model using a 10-fold cross-validation strategy [15]. An accuracy greater than 84 % was shown by all the models involved in the study. The obtained results included a recall of 99.42 %, an accuracy of 86.66 %, and an AUC score of 0.625. The CNN-LSTM model provided the best results with an accuracy of 92.3 %, a recall of 93.68 %, and an AUC score of 0.90 using Holdout Validation.
A total of 4 architectures were studied by Ibrahim et al. For detecting and diagnosing disorders affecting the human lungs [16]. Detection was among three classes, namely, Pneumonia, Lung Cancer, and COVID-19. Out of all the models used for the study, the CNN + VGG19 model performed best yielding an accuracy of 98.05 %. An accuracy of 96.09 % was obtained using GRU + ResNet152V2.
A study to detect abnormalities from chest CT scan images of Pneumonia and COVID-19 patients [17] performed by Ni, Q., et al. aimed at comparing various deep learning models for the task. Results show that our results are superior compared to people who have the expertise in the identification and detection of lesions. According to Xu’s study, the results achieved by their model had a specificity of 67 %, a sensitivity of 74 %, and a total accuracy of 73 %. This study revolved around observing inception-migration-learning models and their performance for the task of differentiating COVID-19 from other infections caused by pathogens such as bacteria, protozoa, viruses, etc.
In another study, Ibrahim, et al. analyzed deep neural network models and their performance using transfer learning for classification between three classes, viz., COVID-19 Pneumonia, Non-COVID-19 Viral Pneumonia, and Bacterial Pneumonia [18]. For multi-class classification, the model obtained 98.19 % sensitivity, 95.78 % specificity, and 94.43 % accuracy. For binary classification among Healthy and Bacterial Pneumonia classes, the model obtained 91.49 % sensitivity, 100 % specificity, and 91.43 % accuracy. For binary classification between COVID-19 Pneumonia and Non-COVID-19 Viral Pneumonia, a testing accuracy of 99.62 % was achieved. Similarly, for classification between COVID-19 Pneumonia and Healthy CXRs, a testing accuracy of 99.16 % was achieved. Classification across Bacterial pneumonia, COVID-19 and Healthy yielded a testing accuracy of 94.00 % and 93.42 % for classification among four classes; Healthy, Bacterial Pneumonia, COVID-19, and non-COVID-19 Viral Pneumonia.
A new transfer learning pipeline consisting of DenseNet-121 and the ResNet-50 networks, called DenResCov-19 [19] was created by Mamalakis, M. et al. This was primarily created for the classification and detection of Pneumonia, COVID-19, Tuberculosis, or Normal using CXRs. The results achieved by the model for classification of Pneumonia, COVID-19, and Normal included an AUC score of 96.51 %, F1 score of 87.29 %, precision of 85.28 %, and an overall recall of 89.38 %.
Gouda et al. considered Deep Learning strategies to predict COVID-19. The study proposed two DL approaches based on ResNet-50 neural network using chest X-ray (CXR) images. COVID-19 Image Data Collection (IDC) and CXR Images (Pneumonia) were used as dataset for the following. The pre-processing was done using augmentation, normalization, enhancement and resizing of the images. To carry out the task, multiple runs of modified version of Resnet-50 was made done to classify the images. The ResNet-50 feature extraction is done by several convolutional and pooling layers. A fully connected and soft-max layer does the classification. The weight and bias values of convolutional and fully commenced layers are tuned using the training algorithm. This training algorithm includes many hyperparameters, which helps to improve the performance of the ResNet-50 model [20]. In terms of performance, the values exceed 99.63 % in many metrics including, F1-score, accuracy, recall, precision and AUC [21]. Mahesh Gour et al. designed a new stacked CNN model for COVID-19 detection. The dataset includes CT images and combination of three publicly available X-ray images. They firstly used different sub -models obtained from VGG-19 and Xception models during the training. Then these were together stacked as softmax classifier. To detect COVID-19 from radiological image data, a stacked CNN model is proposed, combining the differences between CNN sub-models. The sensitivity for Binary classification and Multiclass classification was 98.31 % and 97.62 % respectively[22].
Mahesh Gour et al. developed an automated COVID19 detection model and was named Uncertainty-Aware Convolutional Neural Network Model (UA-CovNet). The model works on the principles of EfficientNet-B3 to fine tune the X-ray images and Monte Carlo dropouts for M passes to obtain the posterior predictive distribution. The sensitivity of the Binary Classification and Multiclass Classification was 99.30 % and 98.15 % respectively. The G-mean of 99.16 % and 98.02 % was seen for both respectively. [23] Yiting Xie et al. believed working on large medical image dataset is really difficult so they carried out their work using ImageNet, a pre-trained model. The pre-trained model can bring in inefficiencies while working on a single channel image. To counter this, they introduced Inception V3 model on ImageNet after the images were transformed into grayscales. The performance was not found waning, hence concluding that colors do not have critical role to play. It was also seen that that grayscale ImageNet pre-trained models had better performance than the color one while classifying diseases from CXRs. [24].
In the 1960 s, a technique for image and signal processing was devised by Thomas Stockham, Ronald W. Schafer, and Alan V. Oppenheim. This technique involved a non-linear mapping to a different domain where linear filters are applied and then mapped back to the original domain [25]. The technique, called Homomorphic Transformation Filter can be employed to enhance the images. It also increases contrast and homogenizes the brightness throughout the image. It can also be used to remove noise from the image. If we take logarithm of the image intensity, we can separate the components of the image linearly in the frequency domain, which are combined multiplicatively. Multiplicative noise includes variations in illumination within the images and can be reduced by applying filtering techniques in the logarithm domain. We can also equalize the low-frequency and high-frequency components of the image to make the illumination more even. This implies that in order to repress low frequencies and intensify high frequencies, high-pass filtering is used in the log-intensity domain [26].
4. X-ray image dataset
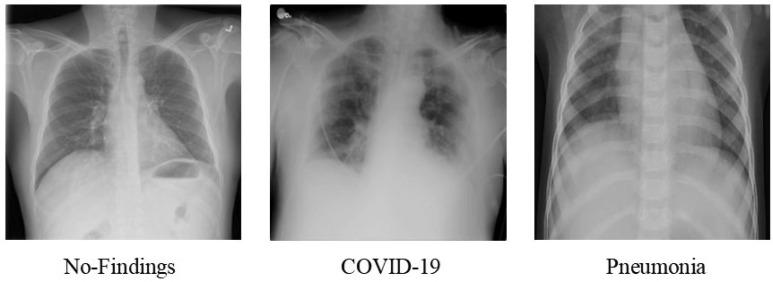
The dataset that we used comprises 2250 images for each of the three classes—COVID-19, Pneumonia, and No-Findings. We use an equal number of images for each class to avoid the problem of class imbalance. Two chest X-ray image datasets are used to achieve our proposed work. The first public dataset is used to extract COVID-19 and No-Findings images and the other public database is used to obtain Pneumonia images. The former database was created in conjunction with medical doctors by researchers from the University of Dhaka, Bangladesh, Qatar University in Doha, Qatar, and colleagues from Malaysia and Pakistan. It makes use of images from 43 different publications as well as the COVID-19 Database of the Italian Society of Medical and Interventional Radiology (SIRM), the Novel Corona Virus 2019 Dataset created by Joseph Paul Cohen, Lan Dao' and Paul Morrison's repository in GitHub [27], [28]. Kang Zhang, Daniel Kermany, and Michael Goldbaum's “Labeled Optical Coherence Tomography (OCT) and Chest X-ray Images for Classification dataset” used CXR images (anterior-posterior) chosen from retrospective cohorts of paediatric patients from Guangzhou Women and Children's Medical Center, Guangzhou, ranging from the age one to five [29]. This dataset was used to obtain CXR images for the class of Pneumonia. To maintain consistency throughout the data used by us, we have resized all of the images to 64 × 64 pixels for further processing. An image of each class obtained from these datasets is shown in Fig. 1 .
Fig. 1.
CXR images from each class, i.e, No Findings, COVID-19, and Pneumonia.
5. Proposed approach
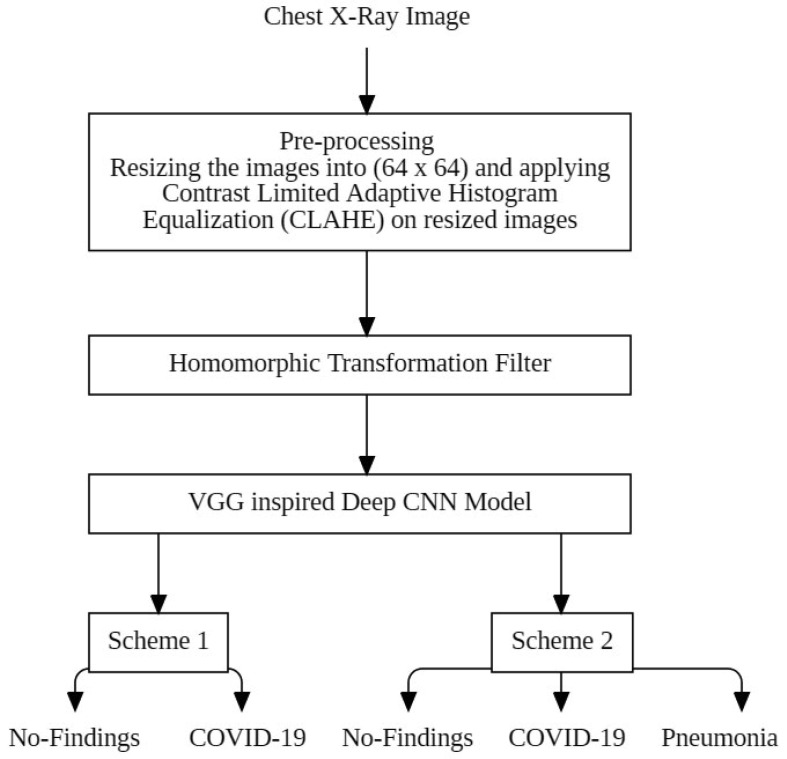
The proposed approach comprises applying pre-processing augmenting methods [30] to our CXR images, including resizing our images to a standard size and applying CLAHE. For a given input image, the algorithm of CLAHE creates non-overlapping contextual regions (also called sub-images, tiles or blocks) and then applies the histogram equalization to each contextual region, clips the original histogram to a specific value and then redistributes the clipped pixels to each gray level [31]. Then Homomorphic Transformation Filter is applied to these processed images. These images are randomly provided to the deep CNN model as input. Deep convolutional neural networks have proven to yield better accuracy when dealing with large volumes of dataset, and many researchers tend to use them as de-facto standards [32]. A typical architecture of CNN consists of multiple blocks with three kinds of layers: convolution, pooling, and fully connected layers [33].The architecture of our deep CNN model is inspired by VGG model’s architecture. Two schemes are employed in order to test the model’s performance. The first scheme comprises binary classification consisting of two classes, COVID-19 and No-Findings. The second scheme involves the classification of CXR images across three classes, namely, COVID-19, No Findings, and Pneumonia. 2250 images have been considered for each class which means scheme 1 involves a total of 4500 images and scheme 2 consists of a total of 6750 images. The block diagram for the suggested model is shown in Fig. 2 .
Fig. 2.
Block Diagram of the suggested model.
5.1. Pre-processing:
Original images taken from both datasets had varied sizes. All of the images were converted into a standard size of 64 × 64 [34] and CLAHE was applied to them. By resizing the images, we can decrease the training time of our model and reduces the memory required for the training purpose. Good thing about having a small size image data is that lot of images can be fed into the model for training without exhausting the memory or increasing the training time. It is a good trade-off between the amount of pixel data in one image and count of images that can be used for training in a limited computational environment. CLAHE helps reduce the noise issue by applying a contrast amplification limiting technique to each neighbouring pixel, which produces a transformation function. To resize the image the resampling using pixel area relation known as INTER_AREA function in OpenCV was used.
Most of recent research studies have used transformation as a key technique for pre-processing of the image data. The idea behind this is to use artifacts derived from a different domain to ease the training process of the model. Typically, the domain is related to either frequency or time. Many different transformation techniques have been used to create the state-of-the-art pre-processing method. Homomorphic Transformation Filter is one such filter belonging to frequency domain that uses Fourier Transformation. This transformation filter has not been explored for image pre-processing in combination with custom CNN for one channel image data (grayscale image) with respect to detection of COVID19. In this research, the combination of CLAHE and Homomorphic Transformation is studied to understand its efficacy as pre-processing techniques. The pre-processing of the image data happens in two stages after the preliminary processing like resizing. First, CLAHE is applied and then Homomorphic Transformation.
5.2. Homomorphic Transformation Filter:
In Homomorphic Transformation Filter, the original domain is nonlinearly mapped to a different domain where linear filtering methods are applied, and then the original domain is mapped back to. A grayscale image can be enhanced via Homomorphic Transformation Filter by simultaneously reducing the intensity range (illumination) and enhancing the contrast (reflection)[35].
| (1) |
Here, = image, l = illumination, r = reflectance.
The equation needs to be converted into the frequency domain in order to be used as a high pass filter. Calculations get more complex because this equation is not anymore, a product equation after the Fourier transformation. In order to help with this problem, natural logarithm is used.
| (2) |
Then, applying Fourier transformation
| (3) |
Or
| (4) |
After that, a high-pass filter on the image is applied which increases the evenness of an image's illumination; the high frequency objects are augmented and the low frequency parts are suppressed.
| (5) |
here, HP = high-pass filter, FI = filtered image in frequency domain.
Then, by using the inverse Fourier transform, frequency domain is returned to spatial domain.
| (6) |
Lastly, to obtain the improved image, we apply the exponential function [35] to remove the log we used earlier.
| (7) |
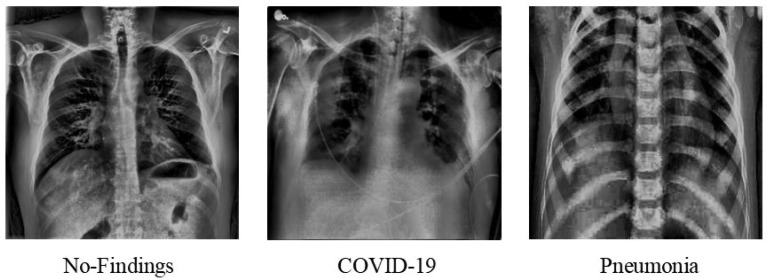
Fig. 3 shows an image from each class after Homomorphic Transformation Filter is applied to the dataset.
Fig. 3.
Chest X-rays after passing through Homomorphic Transformation Filter.
5.3. Grayscale + VGG inspired deep CNN architecture (GrayVIC):
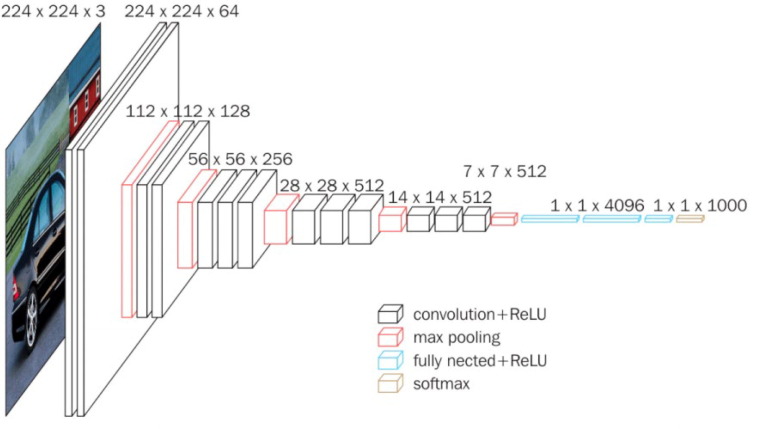
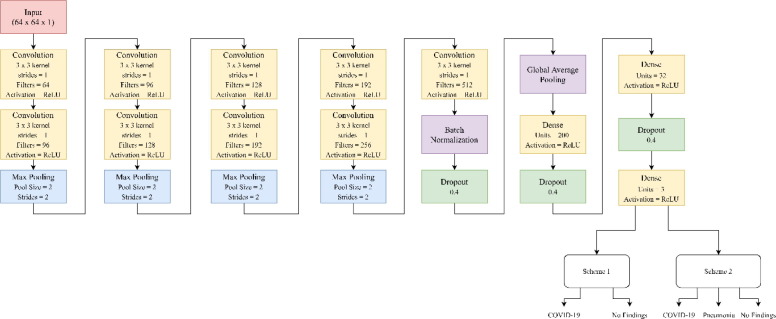
The Convolution Neural Network (CNN) adopted in the proposed work is inspired by VGG models. VGG refers to a typical deep Convolutional Neural Network (CNN) design with numerous layers, and it stands for Visual Geometry Group. The ‘depth’ of a model refers to the number of layers used, with VGG-16 or VGG-19 having 16 or 19 convolutional layers, respectively[36]. In the research domain VGG models are experimented a lot and have mainly been used for transfer learning application, even for Covid-19 detection[37]. In data science, VGG-16 is considered to be one of the most effective classification network whereas VGG-19 is focused more classifying samples effectively [38]. The architecture of a standard VGG-16 model is shown in Fig. 4 , which was used as a reference to build the custom model architecture for this research. Since the proposed model works particularly for grayscale images and is based on VGG style architecture, our model is termed as GrayVIC.
Fig. 4.
Standard VGG-16 architecture.
The proposed model consists of 22 layers (including hidden and dropout layers) and the inputs fed to the model have the shape of 64 × 64 × 1. A sequential model is used where a pattern of one convolutional layer goes after another convolutional layer and then finally a max a max pooling layer is adopted. This same setup is implemented another three times. We then have another convolutional layer followed by batch normalization and dropout. Convolution layers perform feature extraction by convolving the input image with a set of learned kernels. The layer typically consists of a combination of convolution operation and activation function [39]. 2D Global Average Pooling is utilized to flatten the output of previous layers. The mean value of all values over the whole (input width) × (input height) matrix for each of the input channels using a tensor of size (input width) × (input height) × (input channels) is calculated by using the 2D Global Average Pooling block.
This is followed by a dense layer and dropout layer. This is repeated once more successively. A dense layer with softmax as the activation function serves as the final output layer. Except the output layer, all convolutional and dense layers use ReLU as the activation function. ReLU, which stands for rectified linear activation function, is a non-linear or piecewise linear function that, if the input is positive, outputs the input directly; if not, outputs zero. Following the convolution layers is the 2D Max pooling layer. The maximum value for each input channel over an input window of the size specified by pool size is used to down sample the input along its spatial dimensions (height and width). Steps are taken along each dimension to move the window. Max pooling layer estimates the max value of pixel according to filter dimension mentioned in the layer definition. Pooling layer carries out dimensionality reduction by down sampling the values of neurons into a solitary value. Max pooling operation is performed here to combine the output of previous layer into a single value [40].
Before applying Global Average Pooling, we use a batch normalization layer. The model is trained more quickly and steadily using batch normalization. This provides some regularization and helps reduce generalization errors. The dropout layer is applied to the suggested model after the batch normalization layer and dense layers. The regularization is done by the dropout layers which speedup the execution by expelling the neurons whose contribution to the yield is not so high [41]. It also helps us to avoid overfitting the model by ignoring output of some neurons for the upcoming layer [42]. The sum of all inputs is maintained by scaling up non-zero inputs by .
Depending upon the classification type, the number of nodes used in the final output layer is decided. Each of the neurons represents a different class. A softmax function is then used to evaluate this output using the following formula [42],
| (8) |
Here represents the weight vector of the final layer's neuron which represents the output, and v is the fully connected layers’ feature vector before it.
The architecture of the proposed GrayVIC model is shown in Fig. 5 . The suggested model is trained in four scenarios where we use 50 epochs and 100 epochs with and without ReduceLRonPlateau each. When a statistic stops improving, ReduceLRonPlateau lowers the learning rate. Once learning reaches a plateau, models frequently gain by decreasing the learning rate by a factor of 2–10. This scheduler reads a metrics quantity, and the learning rate is decreased if no progress is made after a specified number of “patience” epochs. We have used 0.000001 as our minimum learning rate. This serves as the threshold for our learning rate, implying that our learning rate would not reduce further after the minimum learning rate is encountered.
Fig. 5.
Proposed GrayVIC model architecture.
5.4. Performance analysis
The metrics used in this study are recall, precision, F1-score and accuracy to compare and analyze the performance of our GrayVIC model with other existing models [43]. All of these metrics are obtained from the confusion matrix. We also use AUC score which is obtained from the ROC curve to analyse our model. The likelihood that a random positive example will be placed in front of a random negative example is represented by AUC score. In order to determine the robustness of the model, we employ 5-fold stratified cross validation (CV), which biases the large variations on the test data and averages it on each fold [44] and with CV we also used holdout validation [42] where the training and testing data is split from the total data in a ratio of 4:1, maximizing the data to shape the model [45]. This implies the training data consists of 1800 images for each class and testing data consists of 450 images per class. For the validation set, 10 % of the training data was used to monitor the performance of the model while training. Stratified 5-fold cross validation will ensure the same class ratio throughout the 5 folds as the ratio in the original dataset that is equal counts of each class labels, this removes the tension of class imbalance problem during the training.
We perform multi-class as well as binary classification techniques on our dataset. For binary classification, we classify images between COVID19 and No Findings. For multi-class classification, three classes i.e. No Findings, COVID-19, and Pneumonia are used.
6. Results and discussions
The results of the proposed model are showcased in the following section, with the CXR images transformed using Homomorphic Transformation Filter. The model was compiled based on two classification schemes – binary and multi-class, and two validation schemes – holdout and 5-fold cross validation. 4 different combination pairs of the number of epochs and ReduceLRonPlateau were been tested as part of hyper parameter tuning, apart from tweaking the entire model architecture which has been built from scratch. The public dataset used in this research contains an equal count of images for each class which removes the class imbalance problem. The metrics considered for performance evaluation for the proposed model is Accuracy (ACC), Recall or sensitivity (REC), Precision (PRE), F1 score and lastly AUC value.
6.1. Model performance
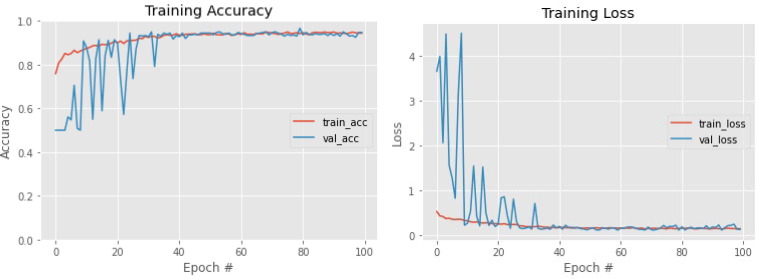
The training and validation accuracy and loss versus epoch plots of the proposed deep CNN model are shown in Fig. 6 . In all the scenarios in which the model was trained, the nature of training accuracy and loss curve were the same. It can be noticed that the model is able to achieve 90 % accuracy within the first 20–25 epochs of the training phase. The ReduceLRonPlateau technique helped in training which can been seen evidently in the graph, it is observed that the validation accuracy/loss fluctuations reduced in the later stages of the training due to lowering of the learning rate by the algorithm which helped in its convergence. The training loss went down to 0.07 in the best fold of cross validation. The training was done in a GPU environment. Total trainable parameter for the proposed model is 2,684,650.
Fig. 6.
Training Accuracy and Training Loss of Proposed Model.
Table 2 portrays the performance of our proposed model when used on multi-class classification on the basis of different combinations of number of epochs and usage of ReduceLRonPlateau technique during the training phase. It is observed that increasing the count of epochs from 50 to 100 is beneficial and the highest accuracy of 0.97 is achieved in the holdout validation. To highlight the robustness of our model and to check that the model is not overfitted, 5-fold stratified cross validation results are used. It shows that the best result is given when the model is trained for 100 epochs with ReduceLRonPlateau. The highest accuracy achieved in 5-fold stratified cross validation is 0.97 and recall is 0.95 along with an AUC value of 0.96. The standard deviation between the fold’s testing accuracies was 0.0121 with an average accuracy of 0.95. The average time taken by the model to train on the dataset consisting of 5400 images for 100 epochs is 8 min 20 s.
Table 2.
Classification performance of proposed GrayVIC model for multiclass classification.
| MODES | MULTICLASS CLASSIFICATION (Hold-out cross-validation) |
MULTICLASS CLASSIFICATION (5-fold cross-validation) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EPOCHS | ReduceLROnPlateau | ACC | PRE | REC | F1 | AUC | ACC | PRE | REC | F1 | AUC |
| 50 | No | 0.95 | 0.94 | 0.93 | 0.93 | 0.95 | 0.86 | 0.90 | 0.85 | 0.84 | 0.89 |
| 50 | Yes | 0.96 | 0.94 | 0.94 | 0.94 | 0.95 | 0.94 | 0.93 | 0.92 | 0.92 | 0.94 |
| 100 | No | 0.97 | 0.95 | 0.95 | 0.95 | 0.96 | 0.95 | 0.94 | 0.93 | 0.93 | 0.94 |
| 100 | Yes | 0.94 | 0.93 | 0.92 | 0.92 | 0.94 | 0.97 | 0.95 | 0.95 | 0.95 | 0.96 |
Table 3 depicts the results obtained from binary classification using our proposed model based on the same methods used for multi-class. As it can be observed, the binary classification’s overall performance exceeds that of multi-class classification task. However, this difference is not very large, unlike in existing research literature. This tells us the proposed model is effective enough for both kinds of classification tasks. The highest accuracy achieved for this task by the proposed model in holdout validation is 0.98. The 5-fold stratified cross validation results shows that the model when trained for 100 epochs and employed with ReduceLRonPlateau technique achieves the highest value across all metrics. The highest cross validation accuracy reached is 0.98 and recall is 0.97 along with the AUC value of 0.97. The standard deviation between the fold’s testing accuracies was 0.014 with an average accuracy of 0.96. The average training time taken by the proposed model to learn from 3600 images for 100 epochs is 5 min and 6 s.
Table 3.
Classification performance of proposed GrayVIC model for binary classification.
| MODES: | BINARY CLASS CLASSIFICATION (Hold-out cross-validation) |
BINARY CLASS CLASSIFICATION (5-fold cross-validation) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EPOCHS | ReduceLROnPlateau | ACC | PRE | REC | F1 | AUC | ACC | PRE | REC | F1 | AUC |
| 50 | No | 0.95 | 0.93 | 0.92 | 0.92 | 0.93 | 0.89 | 0.90 | 0.87 | 0.86 | 0.87 |
| 50 | Yes | 0.96 | 0.94 | 0.94 | 0.94 | 0.94 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 |
| 100 | No | 0.98 | 0.94 | 0.94 | 0.94 | 0.94 | 0.97 | 0.96 | 0.96 | 0.96 | 0.96 |
| 100 | Yes | 0.97 | 0.95 | 0.95 | 0.95 | 0.95 | 0.98 | 0.97 | 0.97 | 0.97 | 0.97 |
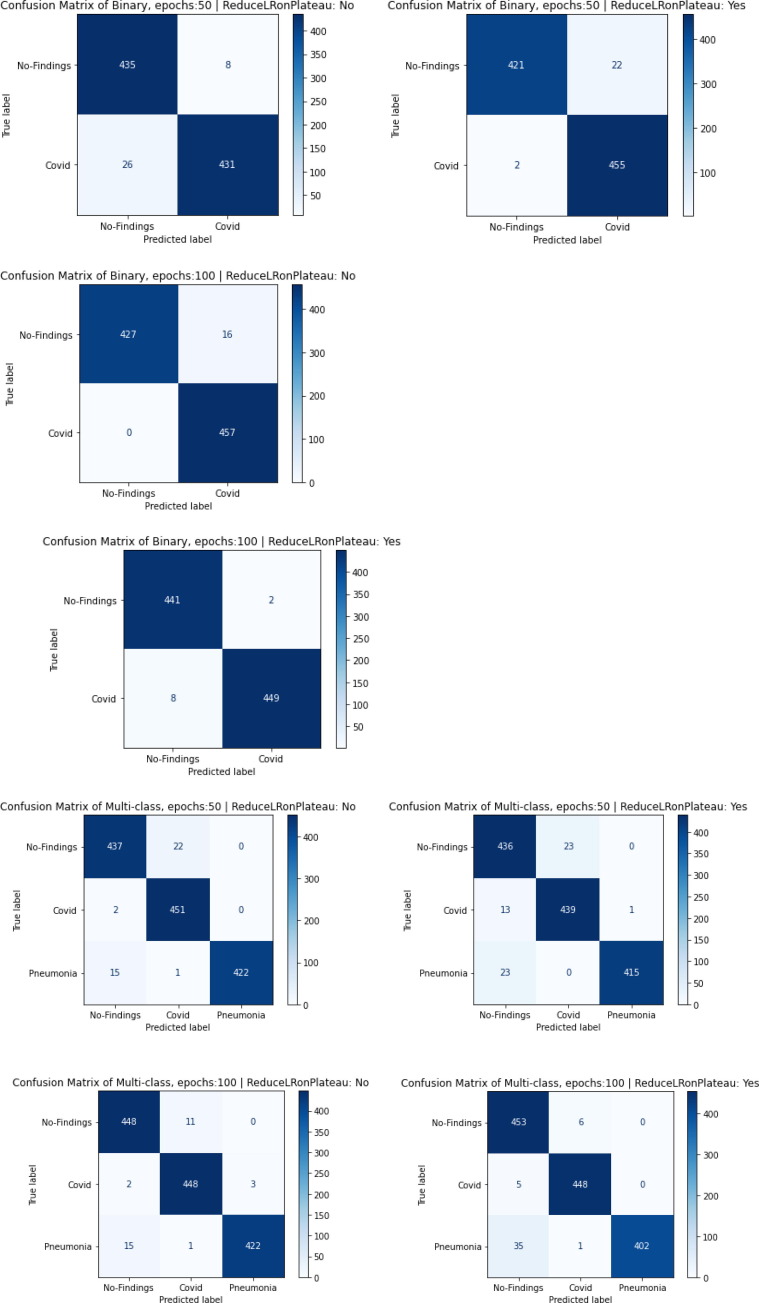
Fig. 7 displays the proposed model’s confusion matrix plots trained with 4 different combinations of hyper parameters. The results belong to the best fold of the 5-fold cross validation of each combination. It has been observed that when the model is trained using ReduceLRonPlateau technique the number of false predictions reduces for the best cases. The ReduceLRonPlateau also reduces the standard deviation to an average value of 0.0049. This helps us ensure that our predictions are close to the average value and these predicted values are spread in a very narrow range. 900 and 1350 images have been used for testing for binary and multi-class classification schemes respectively which ensure that enough instances of each class were used to check the robustness of the model towards each class label. In all the scenarios, it can be observed that the accuracy is above 0.96.
Fig. 7.
The best fold of 5-fold Cross Validation’s confusion matrices for all the 4 cases of model training.
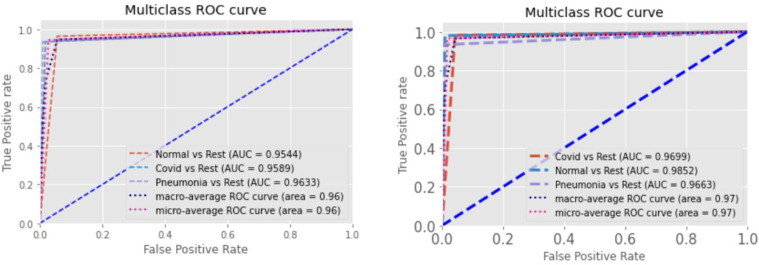
Fig. 8 represents ROC curves from two scenarios. The first ROC curve depicts the values for holdout validation using 100 epochs without ReduceLRonPlateau. The second ROC curve plots the curves for cross-validation using 100 epochs with ReduceLRonPlateau. A plot that shows how well a classification model performs at every level of categorization is called the Receiver Operating Characteristic curve (ROC curve). As visible through the curves, we can see our model is capable of differentiating COVID-19 and the rest of the classes. The values obtained for COVID vs Rest for the first and second curve are 0.96 and 0.97 respectively. Since these values are very close to 1, we can confirm that our model is reliable and robust. The highest AUC score achieved by our model is 0.98 for multi-class classification scheme.
Fig. 8.
ROC curve of the proposed GrayVIC model.
The proposed model’s computational complexity is estimated as follows. When it comes to Deep Learning models, computational complexity plays a critical role. Computational Complexity increases exponentially with the number if network level grown [46]. Computational complexity is often determined with the help of trainable parameters [47] from the model’s architecture. As compared to common transfer learning models, our model projected a better computational result. An approximate total of 2.7 M trainable parameters were required for the proposed model. This is significantly less when compared to 62 M for AlexNet, 25 M for DenseNet, 23.6 M for InceptionV3, 8 M for CapsNet and 4 M for GoogleNet, since only one channel image data is considered.
The model described in the study (as shown in Fig. 5) consists of Conv2D-Conv2D-MaxPool pattern layers which are stacked four times. Batch Normalization layer was used to ease the training of the model and Global Average Pooling layer was used to condense the output of the convolution process to reduce the number of trainable parameters in the final layers of the model. In the proposed architecture, Rectified Linear Activation Unit (RELU) [48] was utilised as an activation function after each convolution layer. The introduction of max-pooling layers reduced computational complexity.
6.2. Model comparison with related works
The proposed work aims to create a VGG inspired CNN model that takes one channel image data as input for detecting and identifying COVID-19 using chest X-rays. All the datasets utilized are treated in accordance with Section 4′s discussion. Hence, the datasets considered for comparative analysis would be different from the dataset considered for the model being proposed.
Most of the papers have used 5-fold cross validation or 10-fold cross validation, and some of them have only done a hold-out validation, this needs to be considered while comparing the results. A precis for the comparative analysis is provided in Table 4 . The comparison is done based on accuracy and recall along with the classification type.
Table 4.
Comparison of existing models to identify and detect COVID-19.
| Ref. No. | Model | Classification Type | Accuracy | Sensitivity/ Recall | CV Type |
|---|---|---|---|---|---|
| Suggested Model | Binary | 98.06 % | 95.12 % | Hold out | |
| Binary | 97.68 % | 96.72 % | 5-fold | ||
| Multi-class | 97.41 % | 94.52 % | Hold out | ||
| Multi-class | 96.56 % | 95.14 % | 5-fold | ||
| [49] | COVID-Caps | Binary | 95.70 % | 90.00 % | Hold out |
| [50] | COVIDX-Net | Binary | 90.00 % | 90.00 % | Hold out |
| [51] | ResNet50 plus SVM | Multi-class | 95.33 % | 95.33 % | Hold out |
| [52] | ResNet-50 | Multi-class | 90.67 % | 96.60 % | Hold out |
| COVID-Net | Multi-class | 93.34 % | 93.30 % | Hold out | |
| [53] | CNN models trained using Transfer Learning | Multi-class | 94.72 % | 98.66 % | 10-Fold |
| [54] | EfficientNet-B0 | Multi-class | 95.24 % | 93.61 % | Hold out |
| EfficientNet-B0 2D curvelet transform |
Multi-class | 96.87 % | 95.68 % | Hold out | |
| [19] | DenResCov-19 | Multi-class | – | 96.51 % | 5-fold |
| DenseNet-121 | Multi-class | – | 93.20 % | 5-fold | |
| [55] | InceptionV3 | Binary | 96.20 % | 97.10 % | 5-fold |
| ResNet 50 | Binary | 96.10 % | 91.80 % | 5-fold | |
| Inception-ResNetV2 | Binary | 94.20 % | 83.50 % | 5-fold | |
| [56] | FractalCovNet | Binary | 98.0 % | 94.0 % | – |
| [57] | Hyperparameter Optimization Based Diagnosis | Multi-class | 88 % | 97 % | – |
| [58] | Multi-task ViT | Multi-class | 85.8 % | 87.43 % | – |
| [22] | Stacked CNN Model | Binary | 97.18 % | 97.42 % | 5-fold |
| [23] | UA-ConvNet | Binary | 99.36 % | 99.30 % | 5-fold |
| UA-ConvNet | Multi-class | 97.67 % |
98.15 % | 5-fold | |
| [59] | BRISK VGG-19 | Multi-class | 96.5 % | 97.6 % | – |
| [60] | 2D- Flexible analytical wavelet transform model (FAWT) | Binary | 93.47 % | 93.6 % | 10-fold |
| [61] | Deep Features and Correlation Features | Binary | 97.87 % | 97.87 % | 5-fold |
| [62] | TL-med Model | Binary | 93.24 % | 91.14 % | – |
| [63] | Cascade VGGCOV19-NET | Binary | 99.84 % | 97.47 % | 5-fold |
| Cascade VGGCOV19-NET | Multi-class | 97.16 % | – | 5-fold | |
| [64] | Inception_ResNet_V2 | Binary | 94.00 % | – | Hold out |
In the first two studies the models described are the COVID-Caps [49] which is capable of handling small datasets and COVIDX-Net [50] which uses seven different architectures. They achieved accuracies of 95.70 % and 90 % respectively for the binary classification scheme. When worked on multi-class classification, with a model ResNet50 combined with SVM [51] attained an accuracy of 95.33 %. This paper advocated that support vector machine, abbreviated as SVM, is quite reliable when compared to other transfer learning models. In [52], the authors compare multiple models for multi-class classification and among the suggested models, COVID-Net performs best with an accuracy of 93.34 % which were trained using hold-out validation. The authors of [53] use transfer learning to extract important information from X-ray images and studied its performance for multi-class classification. They were able to obtain accuracies of 94.72 % and 85 % respectively. A good accuracy of 96.87 % was noticed in [54] using 2D curvelet transform-EfficientNet-B0. This model implemented a blend comprising of chaotic swarm algorithm and two dimensional curvelet transformation.
In [19], the authors have compared DenResCov-19 and DenseNet-121 for X-ray images. Although the accuracies of these models have not been mentioned, the recall of DenResCov-19 is 96.51 % for multi-class classification. The authors of [55] have compared various models for binary classification out of which InceptionV3 produces an accuracy of 96.20 %. The authors of [56] have developed a FractalCovNet architecture for segmentation of chest CT-scan images to localize the lesion region and have trained it using transfer learning for binary classification achieving an accuracy of 98 %. The CNN that was proposed in [57] was a VGG16 which was optimized with five inception modules, 128 neurons in the two fully connected layers, and a learning rate of 0.0027. The proposed method achieved a sensitivity of 97 % for multiclass classification and accuracy it achieved was 88 %.
Multi-task ViT [58] was used for the multiclass which had an accuracy and recall of 85.8 % and 87.43 %. In [22], the authors stacked CNN to create a model which gave the sensitivity score as 97.42 % and accuracy of 97.18. Uncertainty-aware convolution networks were developed in [23] paper which performed well for binary class with an accuracy of 99.36 % and for multiclass it was 97.67 %. Other studies took for comparison shows new variation in deep learning models like the BRISK VGG-19 [59] with 96.5 % accuracy for multiclass whereas for binary type classification there were FAWT model [60], deep and correlation feature model [61]. TL-med model [62] with an accuracy of 93.47 %, 97.8,93.24 % respectively. The Cascade VGGCOV19-Net [63] gave a performance score in terms of accuracy of 97.16 % for multiclass and 99.84 % for binary classification task.
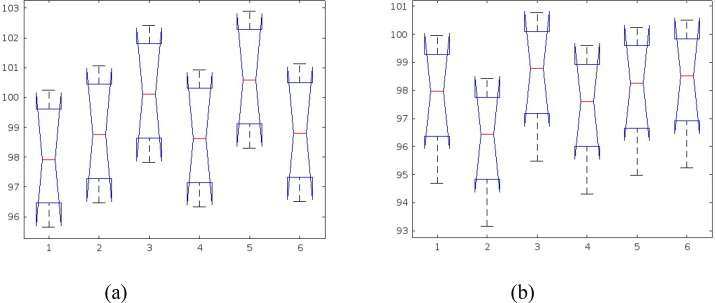
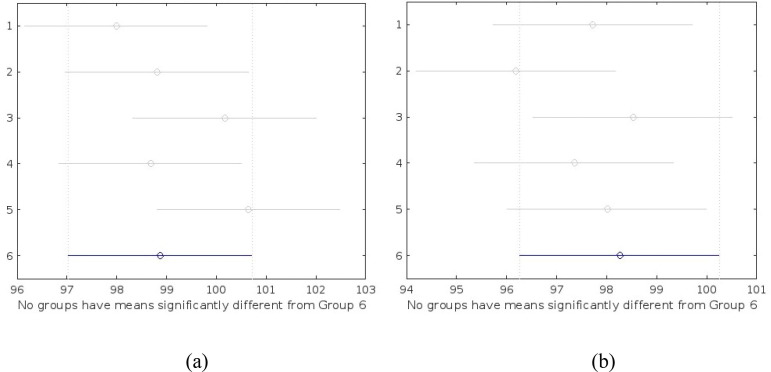
The Analysis of Variance (ANOVA) test was done to evaluate the statistical significance of the results attained from the proposed model. This test was used to infer whether there is a significant difference in the performance of the proposed model along with other related works. The null hypothesis in ANOVA is that there is no difference in means of samples considered for the test. For this test, metric of the top five performing models from the Table 4 is used to analyse the statistical significance of the proposed model, for both binary and multiclass classification task. Tukey's honestly significant difference test (Tukey's HSD) was used to test differences among sample means of proposed models with other related models to estimate the significances. Fig. 9 shows the ANOVA test result graph and Fig. 10 shows the Tukey HSD test result graph for the comparison between the proposed model and the other models. ‘Group 6′ in both graphs denotes the proposed GrayVIC model and other groups denote the models used for comparison purpose.
Fig. 9.
ANOVA test result graph: (a) binary classification; (b) multiclass classification.
Fig. 10.
Tukey HSD test result graph: (a) binary classification; (b) multiclass classification.
Table 5 shows the ANOVA results of binary classification task. From the table it is observed that the p value (0.2541) is more than 0.05 which implies that there is no significant difference in the classification results of the models used for comparison with the proposed model. This means that null hypothesis (H0) is accepted. Tukey HSD test was carried out which showed that the proposed model’s classification result showed a no significant difference in means from the top five performing model used for the statistical analysis test. On a quantitative basis, the proposed model gave better accuracy score than the three of the top five best performing model from the comparison table.
Table 5.
Summary of ANOVA test for binary classification task.
| ANOVA Table | |||||
|---|---|---|---|---|---|
| Source | SS | DF | MS | F | Prob > F |
| Columns | 25.194 | 5 | 5.03875 | 1.42 | 0.2541 |
| Error | 85.383 | 24 | 3.55761 | ||
| Total | 110.576 | 29 | |||
Table 6 shows the ANOVA results of multiclass classification task. From the table it is observed that the p value (0.5335) is greater than 0.05 which implies the classification results of the models used for comparison have no significant difference, thus proving that null hypothesis (H0) is true. This means that the performance of the proposed model is at par with the top five performing models used for the comparison study. Tukey HSD test also pointed towards the same inference and showed no significant difference of the proposed model’s classification result when compared with other top performing models used for the statistical test. The proposed model’s performance was as good as other models with a better computational efficacy.
Table 6.
Summary of ANOVA test for multiclass classification task.
| ANOVA Table | |||||
|---|---|---|---|---|---|
| Source | SS | DF | MS | F | Prob > F |
| Columns | 17.556 | 5 | 3.51113 | 0.84 | 0.5335 |
| Error | 100.121 | 24 | 4.1717 | ||
| Total | 117.676 | 29 | |||
The suggested model has a shorter training time than the other models included in the comparison table since it includes fewer dense as well as convolutional layers. The advantages of the suggested model are:
-
1)
The number of parameters for the proposed model is around 2.7 million which is less than VGG-16 and MobNetV2 architecture.
-
2)
The training time for the model for both classification tasks is approximately 5 to 8 min.
-
3)
The ReduceLRonPlateau technique restricts the fluctuations of validation accuracy during the training of the model.
This model can also be used for feature extraction since fully connected layers can be removed at the end. The objective of this proposed model was to make it work on grayscale images, since it is specifically trained on them, it can be used as feature extraction model that can obtain useful artefacts from grayscale images like X-rays in a more effective manner. The main focus of the suggested model is to help the health care sector in reducing the burden on the medical staff by providing a quick screening system to identify the critical CXR images. The proposed model can also be used for CT scans but it will not be as feasible as CXR images due to its expensive nature and its availability only in large multinational hospitals. In addition to that, it is pointless to conduct expensive CT scans for patients with mild symptoms of asymptomatic nature [34]. In such cases, screening of patients using CXR images will be a much more essential and beneficial method of diagnosis. Additional medical attention can be provided to the patients who are identified by the proposed model as COVID-19 positive. On the other hand, negative cases can be restricted from RTPCR tests to avoid wastage of medical kits. The proposed model can be coupled with IoT and cloud applications to develop a patient monitoring system to curb the spread of the virus.
7. Conclusions
In this research work, a robust deep learning CNN model for the medical image screening of Chest X-rays has been developed using the Homomorphic Transformation Filter along with the 3D-CNN model inspired by VGG architecture for grayscale images. The custom dataset, which was produced from two separate publicly accessible benchmarked datasets, was used to test the model. This custom dataset contains 2250 images for each class (No Finding – Covid – Pneumonia). Two schemes have been used for classification purposes – scheme 1 is a binary classification of COVID-19 from no findings, with a dataset consisting of 4500 images and scheme 2 is a multi-class classification of differentiating COVID-19 from viral pneumonia and no findings, with a dataset consisting of 6750 images. The model had a total of 2.7 M trainable parameters and was trained in a GPU environment. The model has been trained using holdout validation and 5-fold stratified cross validation on the dataset for both classification scheme. The CXRs has been first transformed using Homomorphic Transformation and enhanced using CLAHE. The deep CNN model is trained using this pre-processed output to learn the trainable weights which will enable it to detect the COVID cases. The proposed model successfully classified the COVID-19 cases from Viral-Pneumonia and No-Findings, with precision, recall, F1 score, accuracy, and AUC values of 0.95, 0.95, 0.95, 0.97, 0.96 and 0.97, 0.97, 0.97, 0.98, 0.97 for multi-class classification and binary classification, respectively. Additionally, ReduceLRonPleateau technique is used for curbing the fluctuations in validation accuracy during the training of the model. This ensures that weights are not updated drastically once it reaches near the optimum in the final epochs. To understand the robustness of the proposed model, confusion matrix and ROC curve has been estimated which shows that the model is reliable for the task at hand. Since there are a smaller number of CNN models that takes one channel image data as input, it is difficult the estimate the full potential of this architecture. The proposed model has been compared with the current best approaches used in the research community. The comparative study shows that the proposed works better in all cases and is also efficient due to the model’s simple architecture style. Furthermore, the model was tested using hypothesis test to estimate its statistical significance for both binary and multiclass classification task. The training time of our model is around 8 min for multi-class classification scheme which had a training data of approximately 5,000 images. The model needs to be further tested for its generalization power on new CXRs of COVID-19 cases since a lot of new variants are emerging which can be difficult to be identified by the current model because it was trained on the current publicly available dataset which does not contain CXRs of latest cases.
CRediT authorship contribution statement
Gerosh Shibu George: Visualization, Investigation, Validation, Writing – original draft. Pratyush Raj Mishra: Data curation, Software, Writing – original draft. Panav Sinha: Data curation, Software, Writing – original draft. Manas Ranjan Prusty: Conceptualization, Methodology, Supervision, Validation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors would like to thank the School of Computer Science and Engineering and Centre for Cyber Physical Systems, Vellore Institute of Technology, Chennai for giving the support and encouragement to proceed with the research and produce fruitful results.
References
- 1.Shi Y., Wang G., Cai X.-P., Deng J.-W., Zheng L., Zhu H.-H., et al. An overview of COVID-19. J Zhejiang Univ Sci B. 2020;21:343–360. doi: 10.1631/jzus.B2000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaim S., Chong J.H., Sankaranarayanan V., Harky A. COVID-19 and Multiorgan Response. Curr Probl Cardiol. 2020;45(8):100618. doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader F., Manla Y., Atallah B., Starling R.C. Heart failure and COVID-19. Heart Fail Rev. 2021;26:1–10. doi: 10.1007/s10741-020-10008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raza A., Estepa A., Chan V., Jafar M.S. Acute Renal Failure in Critically Ill COVID-19 Patients With a Focus on the Role of Renal Replacement Therapy: A Review of What We Know So Far. Cureus. 2020;12 doi: 10.7759/cureus.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng G., Zheng K.I., Yan Q.-Q., Rios R.S., Targher G., Byrne C.D., et al. COVID-19 and Liver Dysfunction: Current Insights and Emergent Therapeutic Strategies. J Clin Transl Hepatol. 2020;8(1):1–7. doi: 10.14218/JCTH.2020.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mokhtari T., Hassani F., Ghaffari N., Ebrahimi B., Yarahmadi A., Hassanzadeh G. COVID-19 and multiorgan failure: A narrative review on potential mechanisms. J Mol Histol. 2020;51:613–628. doi: 10.1007/s10735-020-09915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W., Yan F. Patients with RT-PCR-confirmed COVID-19 and Normal Chest CT. Radiology. 2020;295:E3–E. doi: 10.1148/radiol.2020200702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdulkareem M., Petersen S.E. The Promise of AI in Detection, Diagnosis, and Epidemiology for Combating COVID-19: Beyond the Hype. Front Artif Intell. 2021;4 doi: 10.3389/frai.2021.652669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain D.K., Singh T., Saurabh P., Bisen D., Sahu N., Mishra J., et al. Deep Learning-Aided Automated Pneumonia Detection and Classification Using CXR Scans. Comput Intell Neurosci. 2022;2022:e7474304. doi: 10.1155/2022/7474304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh T., Saurabh P., Bisen D., Kane L., Pathak M., Sinha G.R. Ftl-CoV19: A Transfer Learning Approach to Detect COVID-19. Comput Intell Neurosci. 2022;2022:e1953992. doi: 10.1155/2022/1953992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10:257–273. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- 13.Chen X.-W., Lin X. Big Data Deep Learning: Challenges and Perspectives. IEEE Access. 2014;2:514–525. doi: 10.1109/ACCESS.2014.2325029. [DOI] [Google Scholar]
- 14.Aslan M.F., Sabanci K., Durdu A., Unlersen M.F. COVID-19 diagnosis using state-of-the-art CNN architecture features and Bayesian Optimization. Comput Biol Med. 2022;142 doi: 10.1016/j.compbiomed.2022.105244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alakus T.B., Turkoglu I. Comparison of deep learning approaches to predict COVID-19 infection. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim D.M., Elshennawy N.M., Sarhan A.M. Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput Biol Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni Q., Sun Z.Y., Qi L.i., Chen W., Yang Y.i., Wang L.i., et al. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur Radiol. 2020;30(12):6517–6527. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibrahim A.U., Ozsoz M., Serte S., Al-Turjman F., Yakoi P.S. Pneumonia Classification Using Deep Learning from Chest X-ray Images During COVID-19. Cogn Comput. 2021 doi: 10.1007/s12559-020-09787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mamalakis M., Swift A.J., Vorselaars B., Ray S., Weeks S., Ding W., et al. DenResCov-19: A deep transfer learning network for robust automatic classification of COVID-19, pneumonia, and tuberculosis from X-rays. Comput Med Imaging Graph. 2021;94:102008. doi: 10.1016/j.compmedimag.2021.102008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murugan R., Goel T., Mirjalili S., Chakrabartty D.K. WOANet: Whale optimized deep neural network for the classification of COVID-19 from radiography images. Biocybern Biomed Eng. 2021;41:1702–1718. doi: 10.1016/j.bbe.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gouda W., Almurafeh M., Humayun M., Jhanjhi N.Z. Detection of COVID-19 Based on Chest X-rays Using Deep Learning. Healthcare. 2022;10:343. doi: 10.3390/healthcare10020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gour M., Jain S. Automated COVID-19 detection from X-ray and CT images with stacked ensemble convolutional neural network. Biocybern Biomed Eng. 2022;42:27–41. doi: 10.1016/j.bbe.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gour M., Jain S. Uncertainty-aware convolutional neural network for COVID-19 X-ray images classification. Comput Biol Med. 2022;140 doi: 10.1016/j.compbiomed.2021.105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Y, Richmond D. Pre-training on Grayscale ImageNet Improves Medical Image Classification. In: Leal-Taixé L, Roth S, editors. Comput. Vis. – ECCV 2018 Workshop, vol. 11134, Cham: Springer International Publishing; 2019, p. 476–84. https://doi.org/10.1007/978-3-030-11024-6_37.
- 25.Oppenheim A., Schafer R., Stockham T. Nonlinear filtering of multiplied and convolved signals. IEEE Trans Audio Electroacoustics. 1968;16:437–466. doi: 10.1109/TAU.1968.1161990. [DOI] [Google Scholar]
- 26.Madisetti V.K., editor. The Digital Signal Processing Handbook: Digital Signal Processing Fundamentals. CRC Press; Boca Raton: 2017. [DOI] [Google Scholar]
- 27.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., et al. Can AI help in screening Viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 28.Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Abul Kashem S.B., et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput Biol Med. 2021;132:104319. doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kermany D, Zhang K, Goldbaum M. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification 2018;2. https://doi.org/10.17632/rscbjbr9sj.2.
- 30.Joshi R.C., Yadav S., Pathak V.K., Malhotra H.S., Khokhar H.V.S., Parihar A., et al. A deep learning-based COVID-19 automatic diagnostic framework using chest X-ray images. Biocybern Biomed Eng. 2021;41(1):239–254. doi: 10.1016/j.bbe.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siracusano G., La Corte A., Gaeta M., Cicero G., Chiappini M., Finocchio G. Pipeline for Advanced Contrast Enhancement (PACE) of Chest X-ray in Evaluating COVID-19 Patients by Combining Bidimensional Empirical Mode Decomposition and Contrast Limited Adaptive Histogram Equalization (CLAHE) Sustainability. 2020;12:8573. doi: 10.3390/su12208573. [DOI] [Google Scholar]
- 32.Victor Ikechukwu A., Murali S., Deepu R., Shivamurthy R.C. ResNet-50 vs VGG-19 vs training from scratch: A comparative analysis of the segmentation and classification of Pneumonia from chest X-ray images. Glob Transit Proc. 2021;2:375–381. doi: 10.1016/j.gltp.2021.08.027. [DOI] [Google Scholar]
- 33.Bashar A., Latif G., Ben Brahim G., Mohammad N., Alghazo J. COVID-19 Pneumonia Detection Using Optimized Deep Learning Techniques. Diagn Basel Switz. 2021;11:1972. doi: 10.3390/diagnostics11111972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra A.U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez R.C., Woods R.E. Pearson; New York, NY: 2018. Digital image processing. [Google Scholar]
- 36.Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition 2015. https://doi.org/10.48550/arXiv.1409.1556.
- 37.Namani S, Akkapeddi L, Bantu S. Performance Analysis of VGG-19 Deep Learning Model for COVID-19 Detection, 2022. https://doi.org/10.23919/INDIACom54597.2022.9763177.
- 38.Hamwi W.A., Almustafa M.M. Development and integration of VGG and dense transfer-learning systems supported with diverse lung images for discovery of the Coronavirus identity. Inform Med Unlocked. 2022;32 doi: 10.1016/j.imu.2022.101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abraham B., Nair M.S. Computer-aided detection of COVID-19 from X-ray images using multi-CNN and Bayesnet classifier. Biocybern Biomed Eng. 2020;40:1436–1445. doi: 10.1016/j.bbe.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perumal V., Narayanan V., Rajasekar S.J.S. Prediction of COVID Criticality Score with Laboratory, Clinical and CT Images using Hybrid Regression Models. Comput Methods Programs Biomed. 2021;209 doi: 10.1016/j.cmpb.2021.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perumal V., Narayanan V., Rajasekar S.J.S. Detection of COVID-19 using CXR and CT images using Transfer Learning and Haralick features. Appl Intell Dordr Neth. 2021;51:341–358. doi: 10.1007/s10489-020-01831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nandini G.S., Kumar A.P.S. K C. Dropout technique for image classification based on extreme learning machine. Glob Transit Proc. 2021;2:111–116. doi: 10.1016/j.gltp.2021.01.015. [DOI] [Google Scholar]
- 43.Prusty M.R., Jayanthi T., Velusamy K. Weighted-SMOTE: A modification to SMOTE for event classification in sodium cooled fast reactors. Prog Nucl Energy. 2017;100:355–364. doi: 10.1016/j.pnucene.2017.07.015. [DOI] [Google Scholar]
- 44.Mishra N.K., Singh P., Joshi S.D. Automated detection of COVID-19 from CT scan using convolutional neural network. Biocybern Biomed Eng. 2021;41:572–588. doi: 10.1016/j.bbe.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baltazar L.R., Manzanillo M.G., Gaudillo J., Viray E.D., Domingo M., Tiangco B., et al. Artificial intelligence on COVID-19 pneumonia detection using chest xray images. PLoS One. 2021;16(10):e0257884. doi: 10.1371/journal.pone.0257884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang L., Wang X. COVID-RDNet: A novel coronavirus pneumonia classification model using the mixed dataset by CT and X-rays images. Biocybern Biomed Eng. 2022;42:977–994. doi: 10.1016/j.bbe.2022.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarv Ahrabi S, Scarpiniti M, Baccarelli E, Momenzadeh A. An Accuracy vs. Complexity Comparison of Deep Learning Architectures for the Detection of COVID-19 Disease. Computation 2021;9:3. https://doi.org/10.3390/computation9010003.
- 48.Mk M.V., Atalla S., Almuraqab N., Moonesar I.A. Detection of COVID-19 Using Deep Learning Techniques and Cost Effectiveness Evaluation: A Survey. Front. Artif Intell. 2022:5. doi: 10.3389/frai.2022.912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemdan EE-D, Shouman MA, Karar ME. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images 2020. https://doi.org/10.48550/arXiv.2003.11055.
- 51.Sethy PK, Behera SK. Detection of Coronavirus Disease (COVID-19) Based on Deep Features 2020. https://doi.org/10.20944/preprints202003.0300.v1.
- 52.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10:19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altan A., Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021;24:1207–1220. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munusamy H., Muthukumar K.J., Gnanaprakasam S., Shanmugakani T.R., Sekar A. FractalCovNet architecture for COVID-19 Chest X-ray image Classification and CT-scan image Segmentation. Biocybern Biomed Eng. 2021;41:1025–1038. doi: 10.1016/j.bbe.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lacerda P., Barros B., Albuquerque C., Conci A. Hyperparameter Optimization for COVID-19 Pneumonia Diagnosis Based on Chest CT. Sensors. 2021;21:2174. doi: 10.3390/s21062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park S., Kim G., Oh Y., Seo J.B., Lee S.M., Kim J.H., et al. Multi-task vision transformer using low-level chest X-ray feature corpus for COVID-19 diagnosis and severity quantification. Med Image Anal. 2022;75:102299. doi: 10.1016/j.media.2021.102299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharyya A., Bhaik D., Kumar S., Thakur P., Sharma R., Pachori R.B. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed Signal Process Control. 2022;71 doi: 10.1016/j.bspc.2021.103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel R.K., Kashyap M. Automated diagnosis of COVID stages from lung CT images using statistical features in 2-dimensional flexible analytic wavelet transform. Biocybern Biomed Eng. 2022;42:829–841. doi: 10.1016/j.bbe.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar R., Arora R., Bansal V., Sahayasheela V.J., Buckchash H., Imran J., et al. Classification of COVID-19 from chest x-ray images using deep features and correlation coefficient. Multimed Tools Appl. 2022;81(19):27631–27655. doi: 10.1007/s11042-022-12500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng J., Tan Z., Yu Y., Wang P., Liu S. TL-med: A Two-stage transfer learning recognition model for medical images of COVID-19. Biocybern Biomed Eng. 2022;42:842–855. doi: 10.1016/j.bbe.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karacı A. VGGCOV19-NET: automatic detection of COVID-19 cases from X-ray images using modified VGG19 CNN architecture and YOLO algorithm. Neural Comput Appl. 2022;34:8253–8274. doi: 10.1007/s00521-022-06918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta RK, Kunhare N, Pateriya RK, Pathik N. A Deep Neural Network for Detecting Coronavirus Disease Using Chest X-Ray Images: Int J Healthc Inf Syst Inform 2022;17:1–27. https://doi.org/10.4018/IJHISI.20220401.oa1.