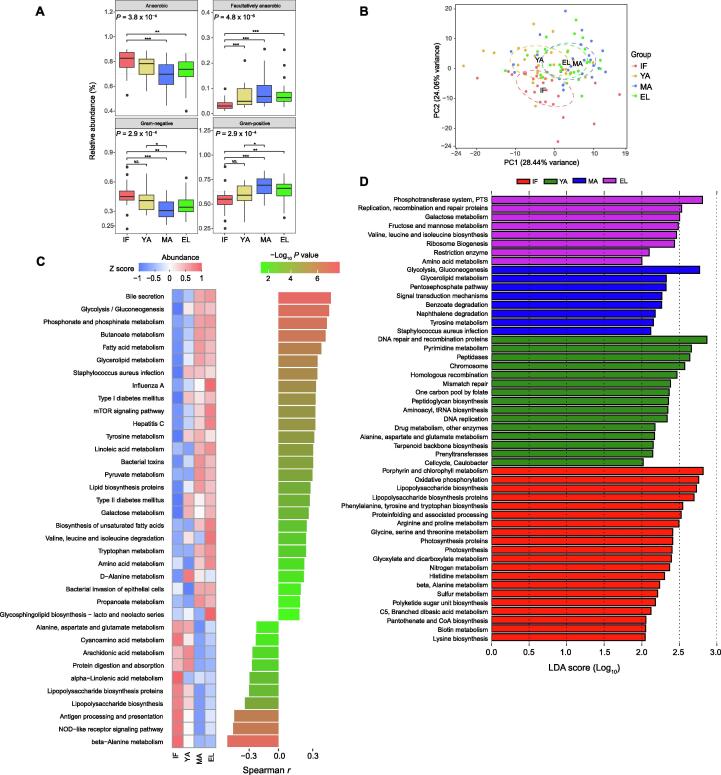
Figure 7.
Age-associated gut microbial phenotypes and functional profiles
A. Comparison of gut microbes with different phenotypes predicted by BugBase among the four age groups. P values for group comparisons are calculated by the nonparametric Kruskal-Wallis test with Tukey’s post-hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS., not significant. B. PCA plot based on microbial function profiles predicted by the PICRUSt software. C. Heatmap illustrating the median abundance and age correlation of gut microbial functions related to the metabolism of carbohydrates, lipids, and proteins as well as host immune response. P values are calculated by the Spearman correlation test. Pathways with FDR < 0.05 are shown. D. LEfSe results of gut microbial functions enhanced in each of the four age groups. PCA, principal component analysis; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.