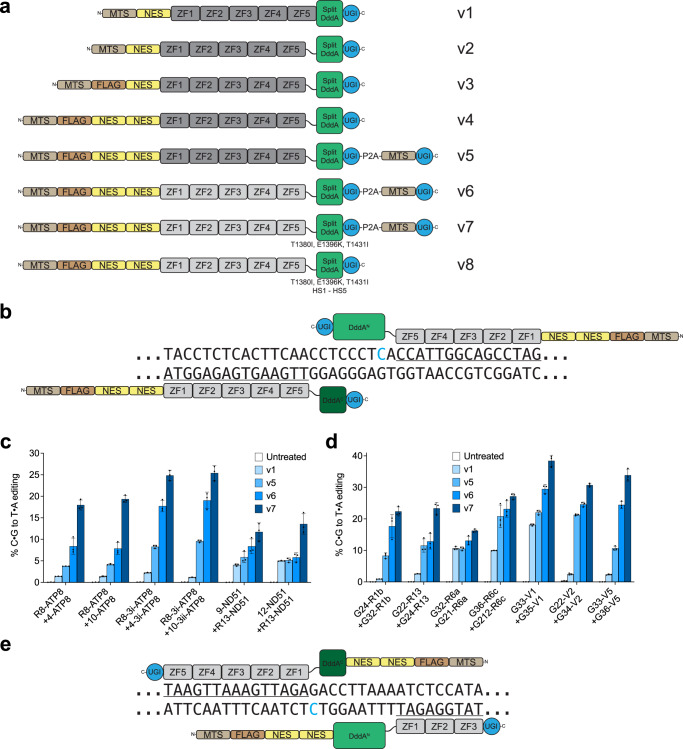
Fig. 1. Optimizing ZF-DdCBEs increases base editing efficiency in mitochondria.
a Architectures of optimized ZF-DdCBEs showing progression from v1 to v8. The components are a mitochondrial targeting signal, FLAG tag, nuclear export signal(s), ZF array with either canonical ZF scaffold (dark grey) or optimized ZF scaffold (light grey), Gly/Ser-rich flexible linker, split DddA deaminase (with or without activity-enhancing mutations and specificity-enhancing mutations) and UGI. b A v8 ZF-DdCBE pair with canonical C-terminal architecture. The ZF-DdCBE pair shown is 9-ND51+R13-ND51. c, d Mitochondrial DNA base editing efficiencies of HEK293T cells treated with (c) six optimized ZF-DdCBE pairs used to establish architectural improvements or (d) seven additional optimized ZF-DdCBE pairs. e A v8 ZF-DdCBE pair with N-terminal architecture. The ZF-DdCBE pair shown is LT51-Mt-tk+RB38-Mt-tk. For (b, e) ZF binding sites are underlined and the cytosine with the highest editing efficiency is colored in blue. For (c–d) values and errors reflect the mean ± s.d. of n = 3 independent biological replicates. The editing efficiencies shown are for the most efficiently edited C•G within the spacing region. Source data are provided as a Source Data file.