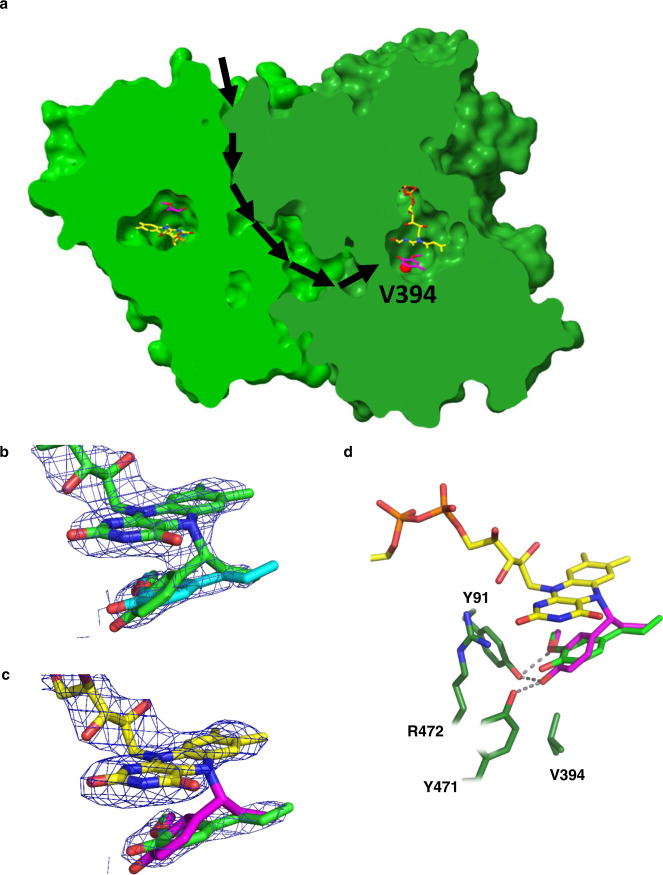
Fig. 5. Structural features of S394V-EUGO5X.
a Position of V394 in the cavity, showing a sliced view of S394V-EUGO5X. The mutated V394, shown as a red sphere, is in close contact with a deep passageway buried between the two chains of the dimer. b, c Detailed views of the electron densities of 4-n-propylguaiacol bound to two S394V-EUGO5X subunits. b unbiased 2Fo-Fc electron density map showing the presence of a stretch of electron density extending from the flavin N5 atom and suggesting the presence of a covalent bond between the flavin and 4-n-propylguaiacol in subunit B. c unbiased 2Fo-Fc electron density map of a non-covalently bound ligand observed in chain A. The maps are contoured at 1.2 σ level and were calculated before the inclusion of the ligands in the refinement. In both panels, the non-covalent and covalent ligands are superimposed. Because of different crystal contacts, the crystalline protein chains might differ with regard to the rates of catalysis, substrate diffusion, or product release explaining why the covalently bound ligand accumulates preferentially in certain protein subunits. d Superposition of the covalent (in magenta) and non-covalent (in green) 4-n-propylguaiacol complexes as observed in two crystallographically independent subunits.