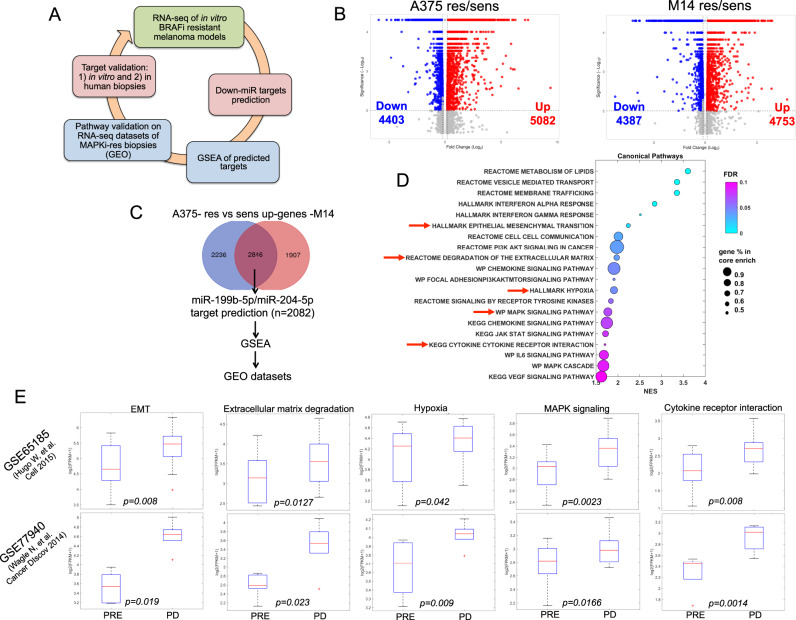
Fig. 2. Core escape pathways of MAPKi resistance in melanoma are controlled by miR-204-5p and miR-199b-5p.
A Schematic illustration of the experimental approach to identify the molecular pathways/targets regulated by oncosuppressive miRNAs impacting MAPKi resistance. B Volcano plots of differentially expressed genes (DEGs) coming from bulk RNA sequencing (RNA-seq) of A375 and M14 BRAFi-resistant cells vs. sensitive counterparts (Log2Fold Change>0.15 and adjusted p value < 0.05). C Venn Diagram of the commonly up-regulated genes of res/sens cells potentially targeted by miR-204-5p and/or miR-199b-5p. D Bubble plot of Gene Set Enrichment Analysis (GSEA) showing 20 enriched gene expression signatures of BRAFi resistant cells. E Box plots representing 5 pathways enriched in biopsies of therapy resistant patients (datasets of bulk RNA-seq numbers GSE65185 and GSE77940): epithelial to mesenchymal transition (EMT), degradation of the extracellular matrix, hypoxia, MAPK signaling pathway and cytokine-cytokine receptor interaction. GSEA was run in preranked mode using classic as metric and 1000 permutations (FDR < 0.1, p value < 0.05, logfold>0.3).