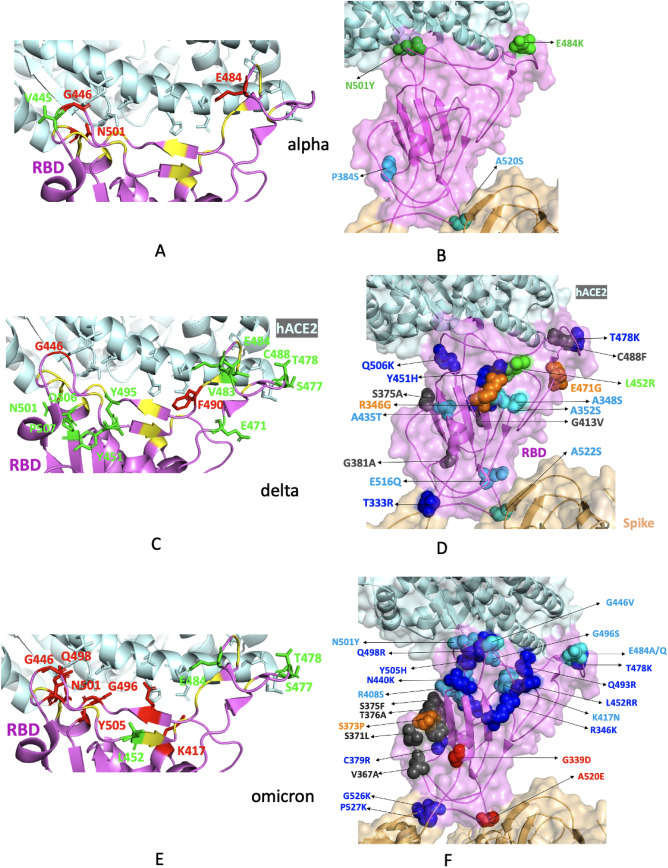
Fig. 3.
Structural mapping of mutations observed in the RBD domain of B.1.1.7 (alpha variant), B.1.617.2 (delta variant) and B.1.1.529 (omicron variant) from India found in the GISAID database from January–May 2021 and December 2021–January 2022. A, C, E RBD mutations in alpha, delta and omicron variants. RBD is coloured violet and hACE-2 is coloured pale cyan. RBD-binding residues of hACE-2 are shown as sticks. The mutations which overlap with the binding region of hACE-2 are coloured red and shown as sticks. The mutations that lie within 5 Å of the hACE-2-binding residues of RBD are coloured green and shown as sticks. Other hACE-2-binding residues are coloured yellow. B, D, F RBD mutations in alpha, delta and omicron variants. RBD is coloured violet, hACE-2 is coloured pale cyan and rest of Spike is coloured brown and shown as a transparent surface. Mutations that exhibit a change in surface charge are shown as spheres and coloured blue and red, polar-to-hydrophobic mutations are coloured grey, hydrophobic to polar mutations are coloured cyan and remaining mutations are coloured orange. The key mutations are coloured green (Color figure online)