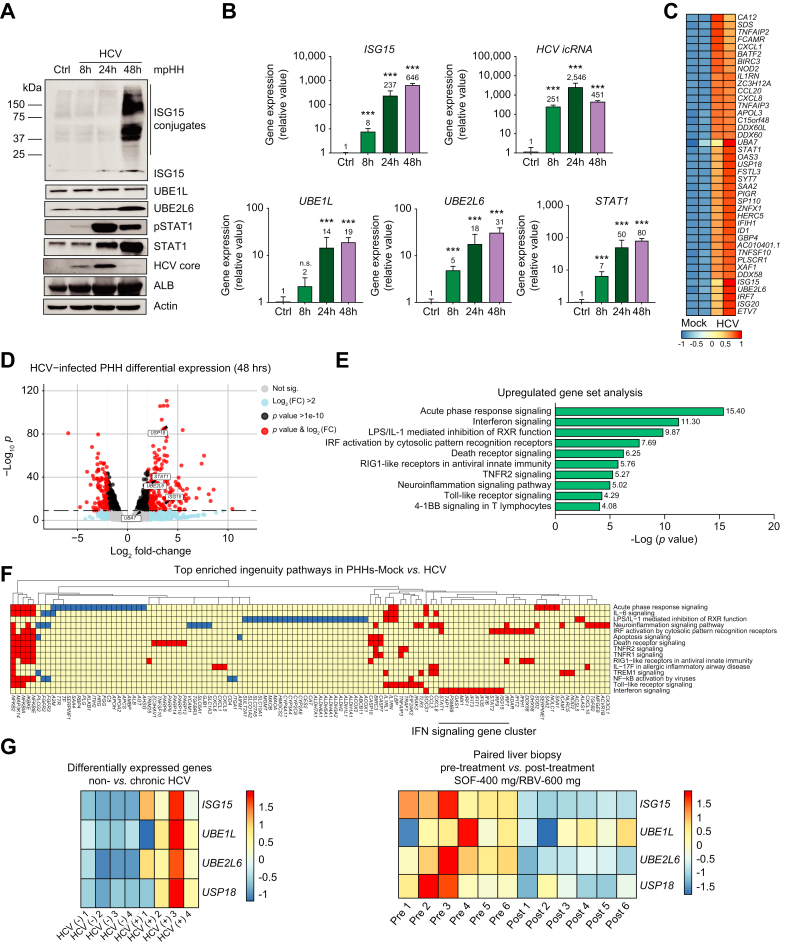
Fig. 1.
ISGylation is detectable in hepatocytes following HCV infection.
(A-B) Mouse-propagated human hepatocytes (mpHHs) were treated with HCV (MOI=1) for 8, 24 and 48 h. (A) Western blot analysis of protein expression for ISG15, UBE1L, UBE2L6, pSTAT1, STAT1, HCV Core Protein and β-actin. (B) qPCR analysis of mRNA expression for ISG15, UBE1L, UBE2L6, STAT1 and HCV intracellular RNA. Data from repeated experiments in triplicate were averaged and are expressed as mean and standard deviation values (error bar) presented with an unpaired Student’s t test with Welch’s correction used to determine the p values. A p value <0.05 was considered significant. ∗p <0.05, ∗∗p <0.001, ∗∗∗p <0.0001, n.s., non-significant. (C-F) RNAseq analysis from PHHs following 48 h of HCV infection (MOI=1) https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE211161 (C) Heat map of gene expression from the top 40 significantly upregulated genes. (D) Volcano plot of gene expression changes stratified by log-fold change and p value. (E) Top 10 upregulated signaling pathways. (F) Top 15 enriched ingenuity pathways. (G-left panel) Heat map of gene expression of ISG15, UBA7, UBE2L6 and USP18 in individuals with and without chronic HCV infection from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84346 published dataset. (G-right panel) Heat map of gene expression of ISG15, UBA7, UBE2L6 and USP18 in humans pre- and post-treatment with sofosbuvir and ribavirin from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51699 published dataset. RNAseq data are from one experiment with two technical replicates. mpHHs, mouse-propagated human hepatocytes.