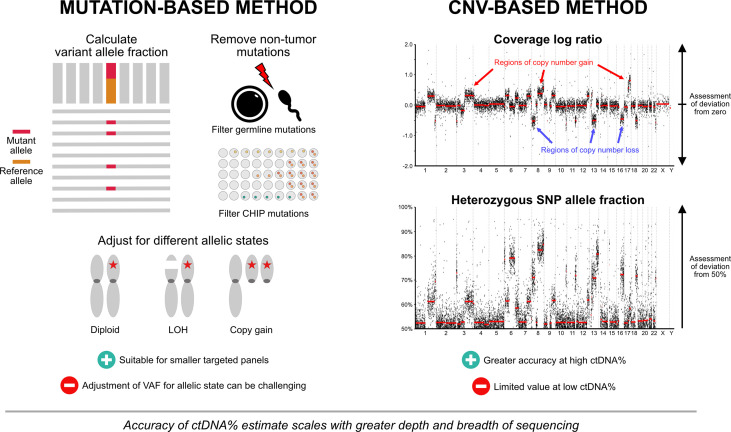
Figure 1.
Approaches to ctDNA fraction estimation. Both mutation-based and copy number-based approaches are used for ctDNA% estimation. Mutation-based approaches typically utilize the variant allele fraction of truncal mutation(s), with or without adjustment for different allelic configurations. Germline mutations and mutations arising from clonal hematopoiesis of indeterminate potential are removed to prevent contribution to ctDNA% estimates. Mutation-based methods can be complemented by systematic assessments of genomic regions affected by copy number changes. Regions of copy gain or loss are identified by evaluating sequencing read depth ratios and/or shifts in B-allele frequency at germline heterozygous SNP loci. The degree of signal from segment-level copy number alterations can then be used with overall tumor ploidy estimates to calculate ctDNA%. Panels that provide both mutation and copy number data encompassing broad genomic territory are likely to yield the most accurate estimates across a spectrum of ctDNA%. Abbreviations: CHIP, clonal hematopoiesis of indeterminate potential; ctDNA%, circulating tumor DNA fraction; LOH, loss-of-heterozygosity; SNP, single nucleotide polymorphism; VAF, variant allele fraction.