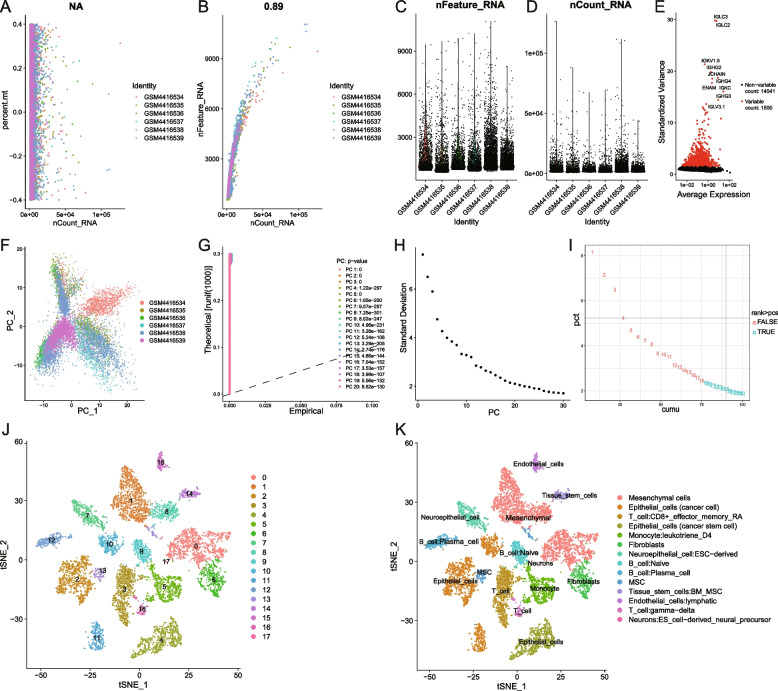
Fig. 2.
scRNA-seq data processing and analysis. A The correlation analysis between sequencing depth and mitochondrial genes using Pearson’s method, NA represents no correlation; B The correlation analysis between the number of genes and sequencing depth using Pearson’s method, the Pearson correlation coefficient was 0.89; C The sequencing depth of 9885 cells from 6 ovarian cancer patients; D The number of genes of 9885 cells from 6 ovarian cancer patients; E Detection of the highly variable genes across the cells in volcano plot, the top 10 genes were marked out; F PCA plot of scRNA-seq samples from 6 patients; G The p values of PCs from 1–20 calculated by JackStraw function; H The standard deviation of 1–30 PCs calculated using ElbowPlot function; I Calculation of the cumulative percentages for each PC, 18 is the last point where change of % of variation is more than 0.1%; H The t-SNE algorithm divided the cells into 18 clusters by 18 PCs; I 18 cell clusters were annotated into 14 cell types