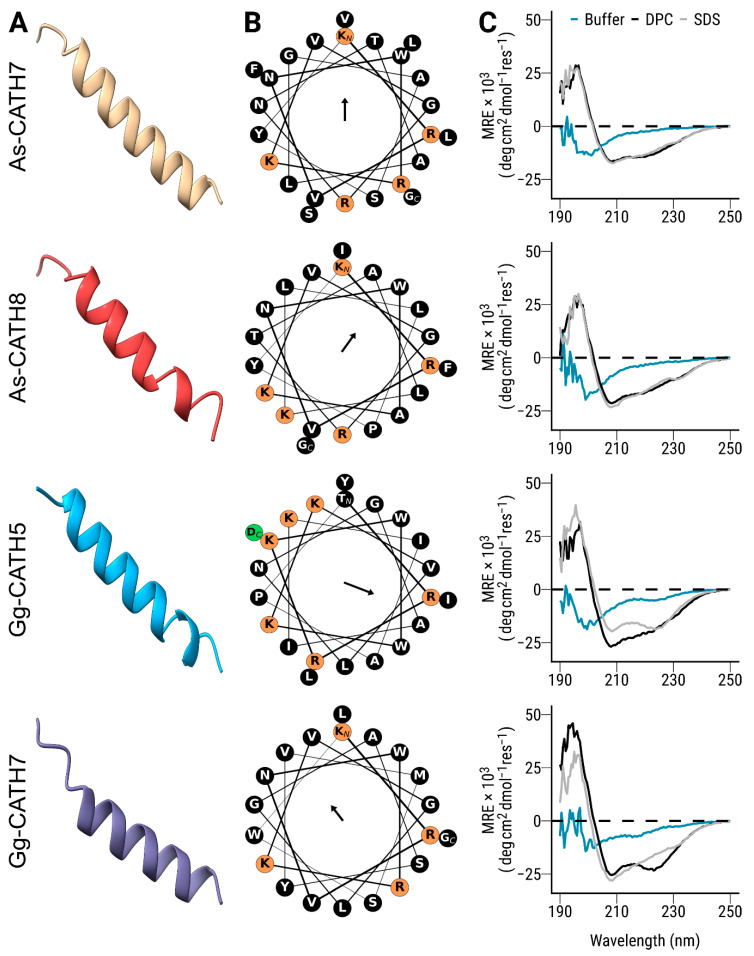
Figure 2.
Structural analysis of the synthetic crocCATHs. (A) Three-dimensional structures were modeled using the AlphaFold algorithm and visualized in Chimera X. (B) Helical wheel representations were generated using the Python package modlAMP. Residues with positive and negative charges are highlighted in orange and green, respectively, whereas the remaining amino acids in the sequence (largely hydrophobic) are shown in black. The orientation of the vector mean hydrophobic moment (amphipathicity) for each sequence is displayed in the center of the wheel. The length of the arrow is proportional to the numerical values shown in Table 1. (C) Circular dichroism spectra of crocCATHs were obtained in sodium phosphate buffer (aqua), as well as in the presence of SDS (grey) and DPC (black) micelles. The results are shown as mean residual ellipticity (MRE).