Fig. 4.
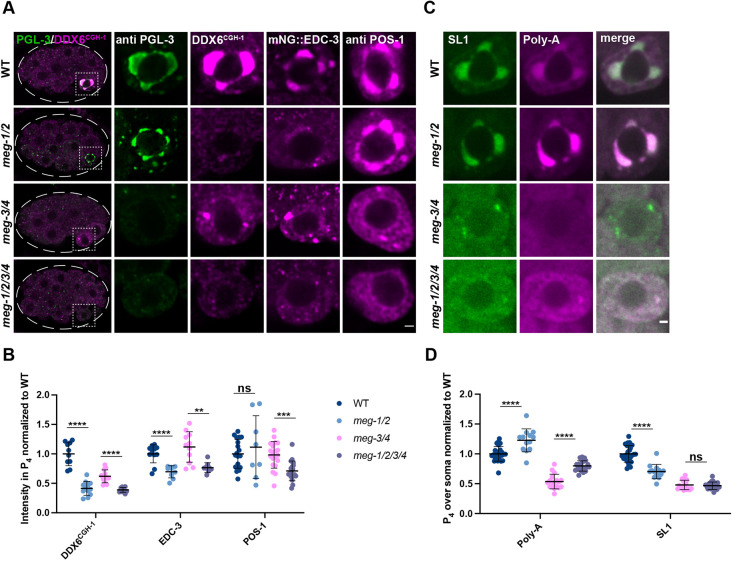
MEG-1/2 are required for maintenance of germline P-bodies in P4. (A) Airyscan photomicrographs of embryos of the indicated meg genotypes co-stained for PGL-3 and CGH-1 (DDX6CGH-1) (whole embryo and P4 inset), or expressing mNG::3×FLAG::EDC-3 and stained for FLAG, or stained for POS-1. meg-1 meg-2 are not essential for localization of PGL-3 or POS-1 to P4 but are required for maintenance of CGH-1 and EDC-3. Dashed white line indicates embryo boundary. Dashed square indicates P4. (B) Intensity of CGH-1, EDC-3 and POS-1 in P4 relative to wild type. Quantification of CGH-1 for each genotype is from one experiment in which mutant and control animals were processed in parallel. Wild type n=10; meg-1/2 n=12; meg-3/4 n=12; meg-1/2/3/4 n=10. Quantification of EDC-3 for each genotype is from one experiment in which mutant and control animals were processed in parallel. Wild type n=12; meg-1/2 n=9; meg-3/4 n=11; meg-1/2/3/4 n=9. Quantification of POS-1 for meg-1 meg-2 embryos is from one experiment and for meg-3 meg-4 and meg-1 meg-2 meg-3 meg-4 from two experiments in which mutant and control animals were processed in parallel. Wild type n=19; meg-1/2 n=8; meg-3/4 n=20; meg-1/2/3/4 n=19. (C) Photomicrographs of P4 in the indicated genotypes probed for SL1 and poly-A. Poly-A levels are increased in meg-1 meg-2 mutants, despite SL1 levels decreasing or not changing. (D) Quantification of poly-A and SL1 in P4 over soma normalized to wild type. Quantification for meg-1 meg-2 embryos is from two experiments and for meg-3 meg-4 and meg-1 meg-2 meg-3 meg-4 from three experiments in which mutant and control animals were processed in parallel. Wild type n=26; meg-1/2 n=13; meg-3/4 n=17; meg-1/2/3/4 n=20. Data are mean±s.d. ****P≤0.0001; ***P≤0.001; **P≤0.01; ns, not significant (unpaired two-tailed t-test). Scale bars: 1 µm.