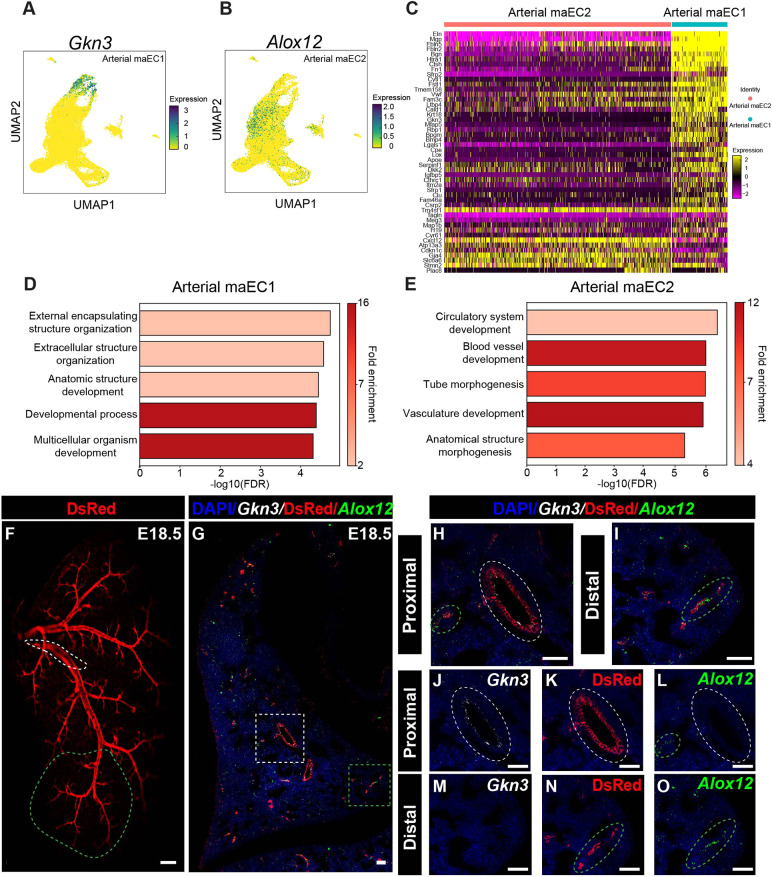
Fig. 3.
Identification of proximal and distal arterial ECs by RNA FISH and predicted function analysis. (A) UMAP embedding of cells colored by Gkn3 expression. (B) UMAP embedding of cells colored by Alox12 expression. (C) Heat map representing expression of hallmark genes in arterial macro endothelial cell types 1 and 2 (arterial maEC1 and maEC2). (D) GO analysis of the maEC1 cluster. (E) GO analysis of the maEC2 cluster. (F) Whole-mount image showing Cxcl12-DsRed arterial endothelium. Dashed white line marks the proximal artery branch and the dashed green line marks the arterial tree from the distal tip of the secondary branch to the quinary branches. (G) RNA FISH for Gkn3 and Alox12 and IHC for DsRed protein at E18.5 proximal (white box) and distal (red box) regions. (H) Higher magnification of RNA FISH for Gkn3 and Alox12 and IHC for DsRed protein in the proximal region. (I) Higher magnification of RNA FISH for Gkn3 and Alox12 and IHC for DsRed protein in the distal region. (J-L) Separate channels for proximal region RNA FISH for Gkn3 (J), IHC staining for DsRed (K) and Alox12 (L). (M-O) Separate channels for proximal region RNA FISH for Gkn3 (M), IHC staining for DsRed (N) and Alox12 (O). Scale bars: 50 mm (F); 50 μm (G-O).