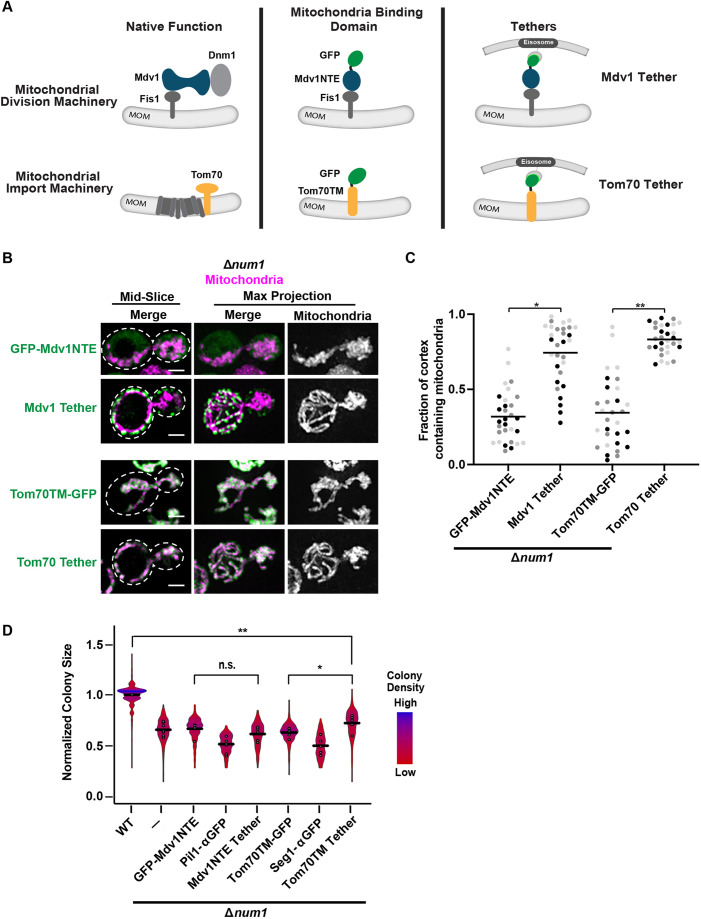
Fig. 3.
Anchoring mitochondria to the cell cortex in the absence of Num1 does not rescue the Δnum1 phenotype. (A) Schematic of Mdv1 and Tom70 in their native complexes and the mitochondrial-binding domains of each protein used in the modified GFP–αGFP nanobody mitochondria-tethering system. MOM, mitochondrial outer membrane. (B,C) Cells expressing mito-dsRED and GFP–Mdv1NTE, Mdv1 Tether, Tom70TM–GFP or Tom70 Tether in the absence of NUM1 were grown in respiratory growth conditions and analyzed by fluorescence microscopy. Whole-cell maximum intensity projections and single focal planes of the middle of each cell are shown (B). Dashed white lines denote the outline of the cell. Scale bars: 2 µm. The graph (C) represents a quantification of the fraction of the cell cortex at mid-cell that is occupied by mitochondria; n=30 cells per strain from three independent experiments, each shown in a different color. Black line denotes the grand mean. (D) Quantification, presented as a violin plot, of WT, Δnum1, Δnum1 GFP-Mdv1NTE, Δnum1 Pil1-αGFP, Δnum1 Mdv1 Tether, Δnum1 Tom70TM-GFP, Δnum1 Seg1-αGFP and Δnum1 Tom70 Tether cell growth at 35°C in respiratory growth conditions as described in Fig. 1E. Black line denotes the grand mean of at least five independent experiments and the circles depict the mean of each independent experiment; n≥316 colonies per strain. Δnum1 and Δnum1 Pil1-αGFP data are recapitulated from Fig. 2B and F, respectively, for comparison. **P≤0.01, *P≤0.05, n.s., not significant (two-tailed unpaired t-test).