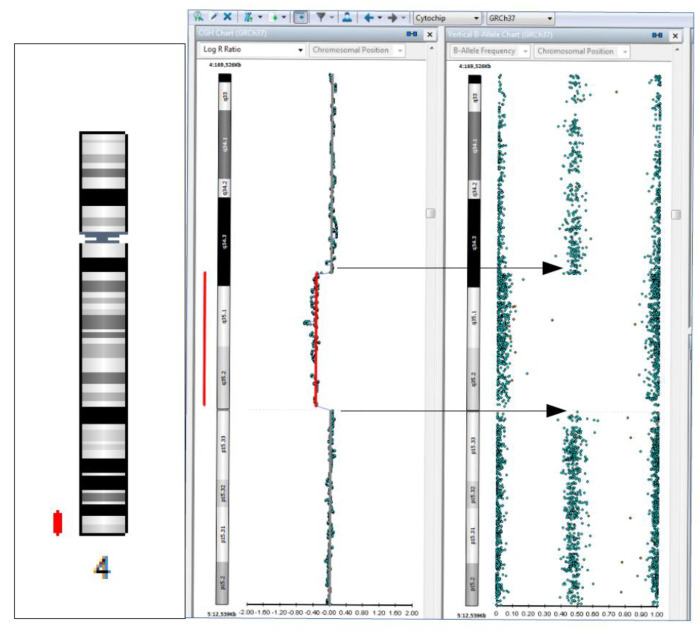
Figure 2.
Deletion of 8.5 Mb (in chromosomal regions marked with red lines), with pathogenic significance, detected in the 4q34.3q35.2 region, containing 21 OMIM genes. The deletions comprising the 4q31q35 region have been known and reported in clinical databases and scientific literature as being responsible for phenotypic manifestations corresponding to the 4q terminal deletion syndrome (or distal monosomy 4q), including craniofacial anomalies, dysmorphic features, intellectual disabilities, developmental delay, ocular, cardiac, genitourinary malformations, and pelvic/limb dysmorphism (ORPHA:96145) [4,5]. Based on all this evidence, both 3p26.3p24.3 duplication and 4q34.3q35.2 deletion detected in the patient’s sample were classified as pathogenic CNVs [4], and contributed to the patient’s phenotypic expression; their simultaneous presence in the case of a single patient has not been reported until now, to our knowledge. The two detected genomic changes could be associated with an unbalanced translocation with a possible parental origin.