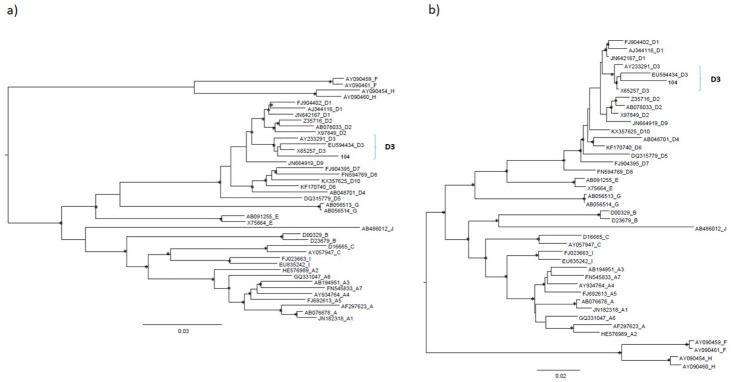
Figure 1.
The maximum likelihood phylogenetic tree built on the first (a) and second dataset of HBV (b). The trees were rooted using the midpoint rooting method. The scale bar at the bottom of the tree represents 0.03 and 0.02 nucleotide substitutions per site respectively for the first and second dataset. An asterisk along the branches represents a SH-aLRT ≥ 80% and UFboot ≥ 95%. Accession numbers of the sequences are indicated in the first part of the tip names followed by HBV genotype/sub-genotype. The accession number for isolate 104 is: OP572234. The HBV sub-genotype D3 cluster was highlighted by brackets.