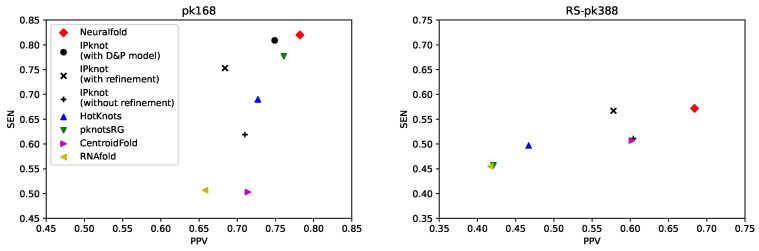
Figure 6.
Positive predictive value–sensitivity (PPV–SEN) plots comparing our algorithm with competitive methods on the pk168 dataset (Left) and the RS-pk388 dataset (Right). For the pk168 dataset, we set , for Neuralfold; , for IPknot with the Dirks–Pierce (D&P) model; , for IPknot with/without refinement; for CentroidFold. For the RS-pk388 dataset, we set , for Neuralfold; , for IPknot without refinement; , for IPknot with refinement; for CentroidFold.