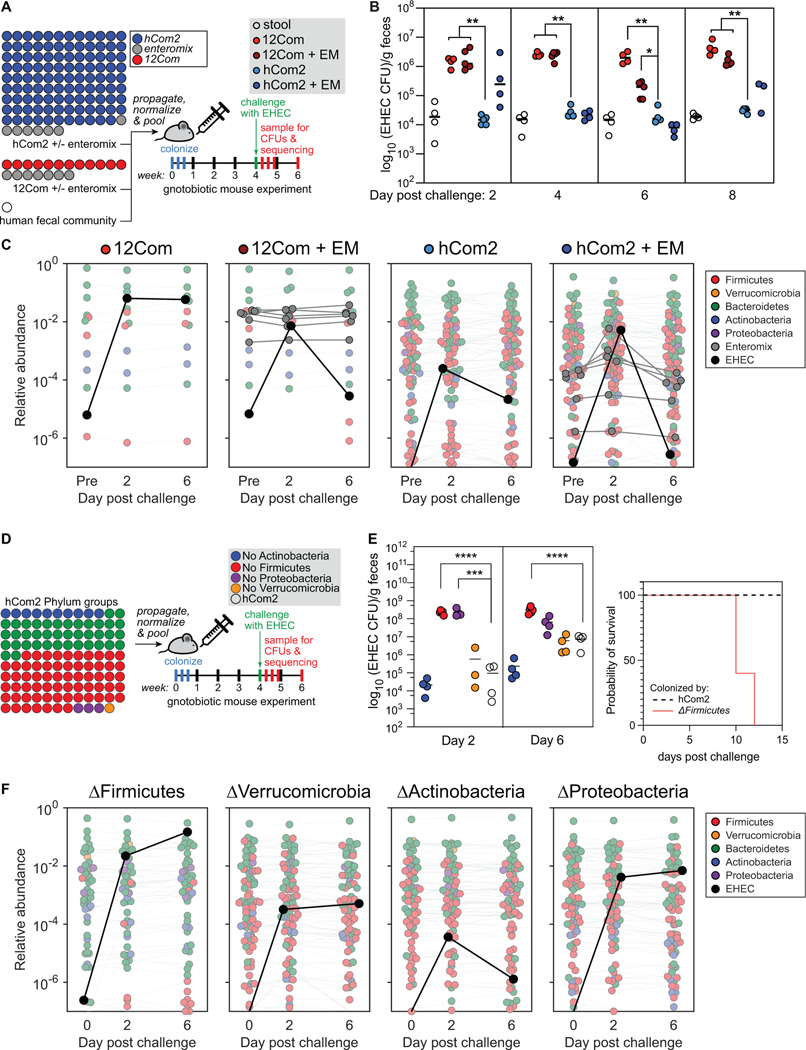
Figure 7: hCom2 exhibits colonization resistance against enterohemorrhagic E. coli.
(A) Schematic of the experiment. We colonized germ-free SW mice with freshly prepared hCom2 or one of two other communities: a 12-member synthetic community (12Com) or a fecal community from a healthy human donor. hCom2 and 12Com do not contain any Enterobacteriaceae; to test whether non-pathogenic Enterobacteriaceae enhance colonization resistance to EHEC, we colonized two additional groups of mice with variants of hCom2 and 12Com to which a mixture of seven non-pathogenic Enterobacteriaceae strains were added (six E. coli and Enterobacter cloacae, Enteromix (EM)). After four weeks, we challenged with 109 colony forming units of EHEC and assessed the degree to which it colonized in two ways: by EHEC-selective plating under aerobic growth conditions, and by metagenomic sequencing with NinjaMap analysis. (B) hCom2 exhibits a similar degree of EHEC resistance to that of a fecal community in mice. Colony forming units of EHEC in mice colonized by the four different communities are shown. As expected, the fecal community conferred robust colonization resistance while 12Com did not. The addition of EM moderately improved the EHEC resistance of 12Com. Despite lacking Enterobacteriaceae, hCom2 exhibited a similar level of EHEC resistance to that of an undefined fecal community. (C) The architecture of hCom2 is stable following EHEC challenge. Each dot is an individual strain; the collection of dots in a column represents the community at a single time point averaged over four co-housed mice. Strains are colored according to their phylum; EHEC is shown in black and members of the Enteromix community are shown in gray. (D) Schematic of the phylum dropout experiment. We colonized germ-free SW mice with four variants of hCom2, each one missing all species from the phyla Actinobacteria, Firmicutes, Proteobacteria, or Verrucomicrobia. After four weeks, we challenged with 109 colony forming units of EHEC and assessed the degree to which it colonized by EHEC-selective plating under aerobic growth conditions, and by metagenomic sequencing with NinjaMap analysis. (E) The ΔActinobacteria and ΔVerrucomicrobia communities retain the ability to resist EHEC invasion, while the ΔFirmicutes and ΔProteobacteria communities are sensitive to EHEC invasion. Right: a large survival difference in ΔFirmicutes-colonized mice compared with hCom2-colonized. (F) The architecture of the phylum dropout communities remains stable following EHEC challenge. Each dot is an individual strain; the collection of dots in a column represents the community at a single time point averaged over four co-housed mice. Strains are colored according to their phylum; EHEC is shown in black.