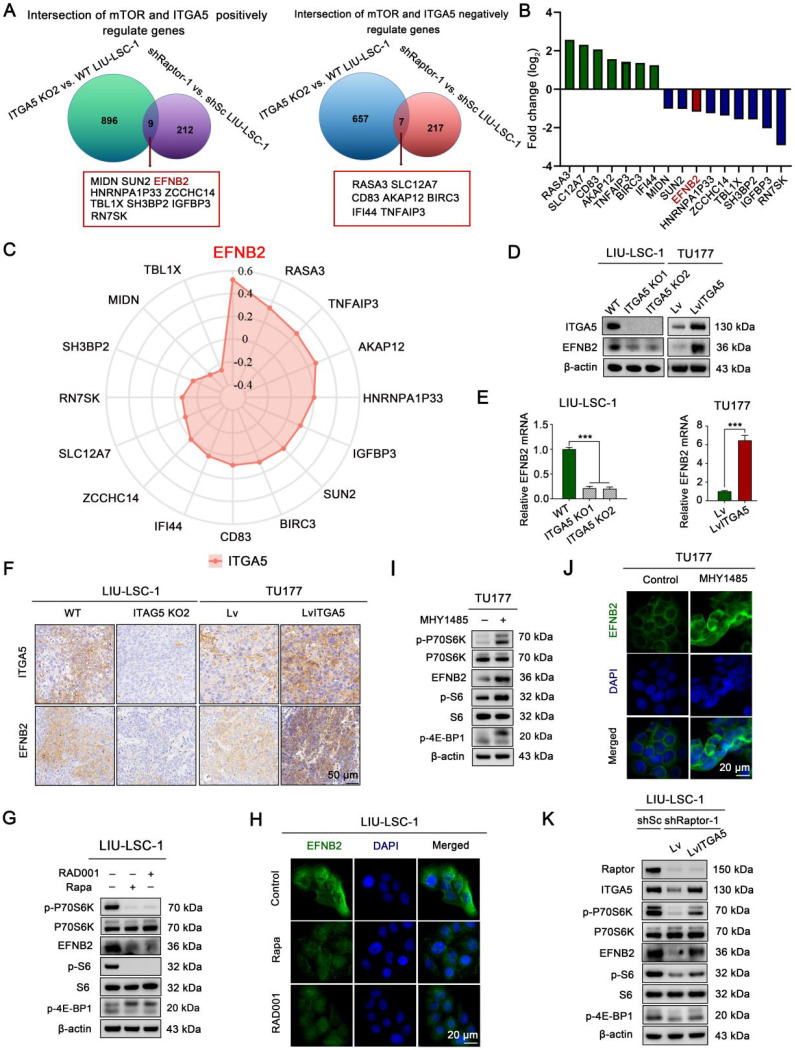
Figure 4.
EFNB2 is a downstream target of the mTORC1-ITGA5 signaling pathway. (A) Schematic illustration for screening of common differential genes (co-DEGs) of the two RNA-sequencing data (ITGA5 KO2 LIU-LSC-1 cells vs. WT LIU-LSC-1 cells and shRaptor-1 LIU-LSC-1 cells vs. shSc LIU-LSC-1 cells). (B) mRNA expression levels of the co-DEGs in the RNA sequencing (ITGA5 KO2 LIU-LSC-1 cells vs. WT LIU-LSC-1 cells). (C) Radar plots were used to demonstrate Pearson's correlations between the expression levels of the co-DEGs and ITGA5 based on TCGA datasets of LSCC. (D-E) EFNB2 expression in the indicated cells was detected by immunoblotting (D) and qRT-PCR (E). Error bars represent the mean ± SD of triplicate technical replicates. ***P < 0.001. (F) Representative IHC images of ITGA5 and EFNB2 staining from the indicated tumor tissues. Scale bars, 50 µm. (G-H) LIU-LSC-1 cells were treated with 20 nM Rapa or 50 nM RAD001 for 24 h. (I-J) TU177 cells were treated with 10 µM MHY1485 for 24 h. (G-J) The samples were detected by western blot analysis (G, I) and IF assay (H, J), scale bars, 20 µm. (K) LIU-LSC-1 cells which endogenous Raptor was abolished by shRNA were infected with lentivirus harboring a vector encoding human ITGA5 or the empty vector. The cell lysates were subjected to western blot analysis.