Figure 6: Pangenome of S. mitis, S. oralis, and S. infantis.
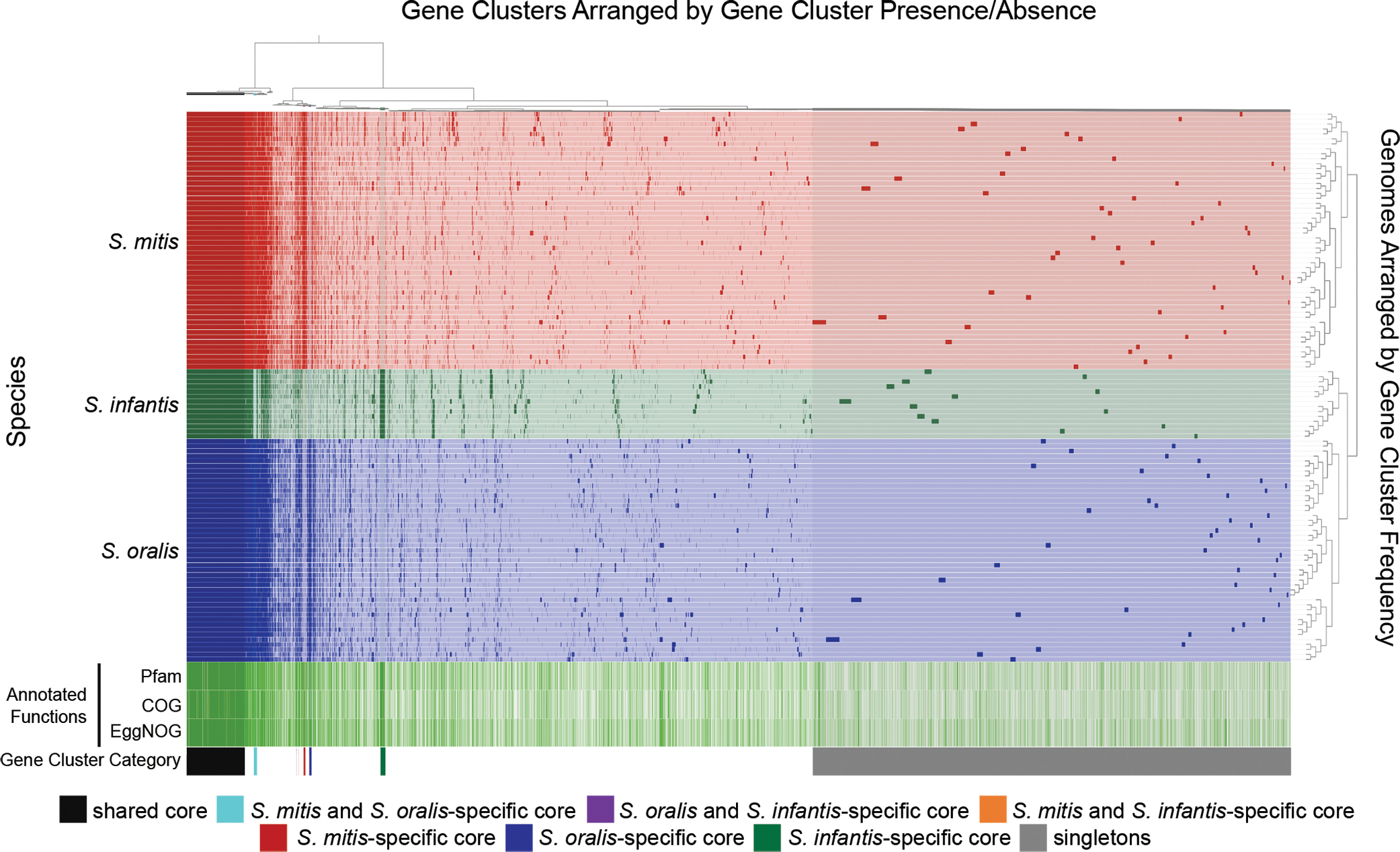
This pangenome is prepared with a more stringent clustering parameter than that of Fig. 5 (see Methods) and shows small blocks of species-specific core genes in addition to a large block of core genes shared among all three species. Genes and genomes are clustered as in Fig. 5. Gene clusters present in all genomes are marked shared core. Gene clusters present in all genomes of one or two species and none of the genomes of the other species are marked as species-specific core. Gene clusters present in a single genome are marked as singletons. The “Annotated Functions” layers indicate whether the gene cluster has (green) or has not (white) been assigned a Pfam, COG, or eggNOG function. In all cases where a species-specific core gene received a functional annotation, genes in the other two species were also assigned that functional annotation, with these exceptions: in S. mitis “PrsW family intramembrane metalloprotease” (Pfam) and “membrane proteinase PrsW, cleaves anti-sigma factor RsiW, M82 family (PrsW)” (COG); in S. oralis "TraX protein” (Pfam); and in S. infantis “UPF0126 domain” and “Competence protein” (Pfam) and “Uncharacterized membrane protein YeiH (YadS) (PDB:5WUC)” and “DNA uptake channel protein ComEC, N-terminal domain (ComEC) or DNA uptake channel protein ComEC C-terminal domain, metallo-beta-lactamase superfamily (ComEC)” (COG). Thus, although species-specific core genes could be detected at the level of protein sequence, functional annotation was similar for all three species.
