Extended Data Fig. 2. Biofilm formation and transfer of mobile genetic elements during interspecies interactions.
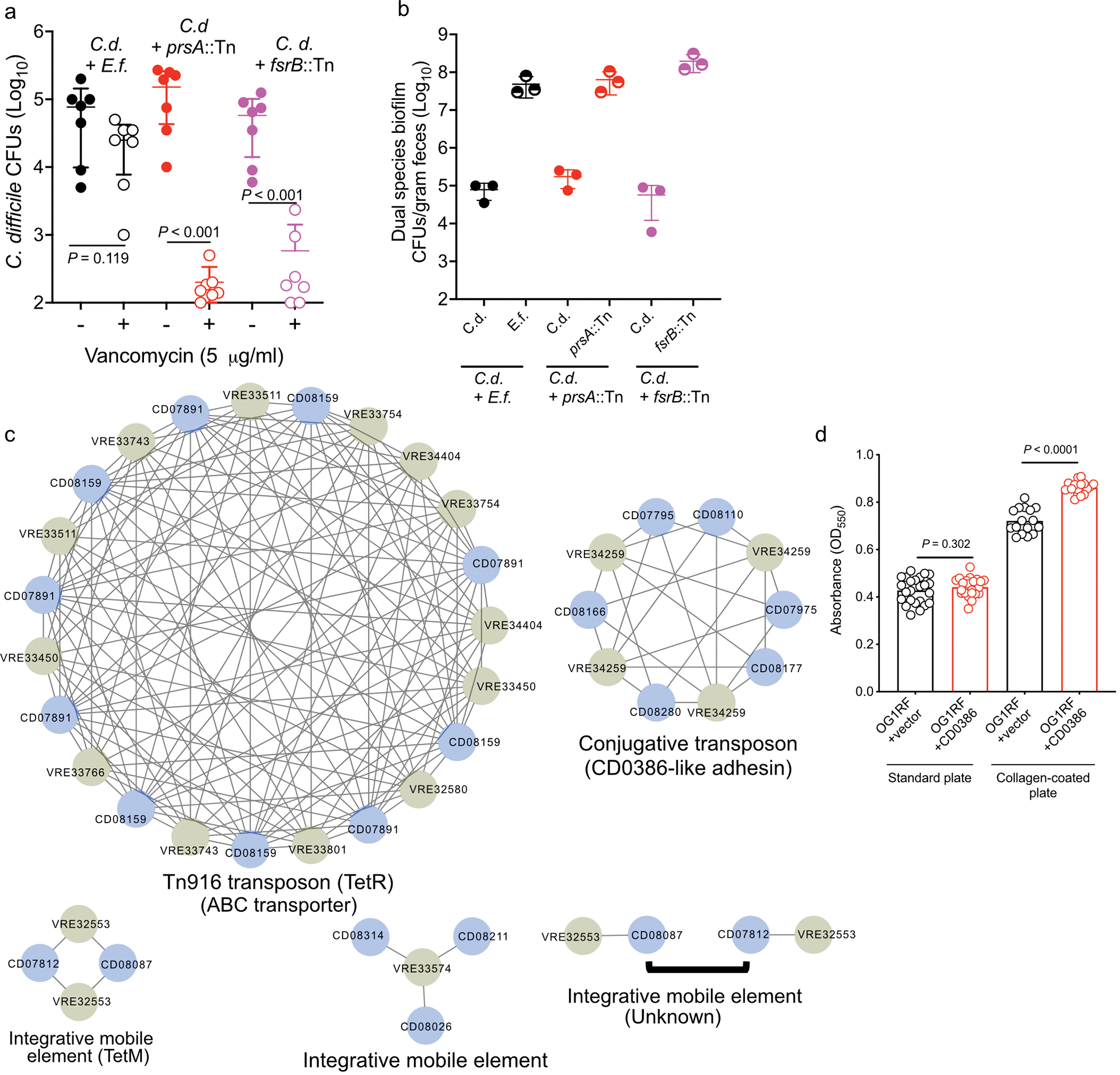
(a) Survival assay of co-culture biofilms with E. faecalis (E.f.) (P=0.119) or transposon mutants in E. faecalis genes OG1RF_11528 (fsrB::Tn) (P<0.001) and OG1RF_10423 (prsA::Tn) (P<0.001). Abundance of C. difficile in untreated (−) or vancomycin treated (+) biofilms are depicted (mean ± s.d., n=7, two-sided Mann-Whitney with Bonferroni-Dunn method for correction for multiple comparisons) (b) Abundance of C. difficile (C.d.) and E. faecalis strains (E.f., prsA::Tn, and frsB::Tn) in untreated dual species biofilms (mean ± s.d., n = 3). (c) Clusters of shared sequences detected in C. difficile (blue) and VRE (olive) genomes of clinical isolates from hospitalized patients. Lines connect sequences with at least 99.98% identity. Clusters are labeled based on mobile element type and relevant cargo, if known. Source data for each cluster can be found in Supplementary Information. (d) Biofilm formation of E. faecalis OG1RF carrying empty pMSP3535 vector or pMSP3535 carrying the CD0386-like adhesin. Biofilm formation was tested in standard (P=0.302) and collagen-coated plates (P<0.001). Crystal violet staining (OD550) values were calculated (mean ± s.d., n = 24/group for standard plates and 16/group for collagen plates, unpaired two-tailed t-tests).
