Extended Data Fig. 5. Transcriptome-guided metabolic flux predictions using genome-scale metabolic network reconstruction for C. difficile.
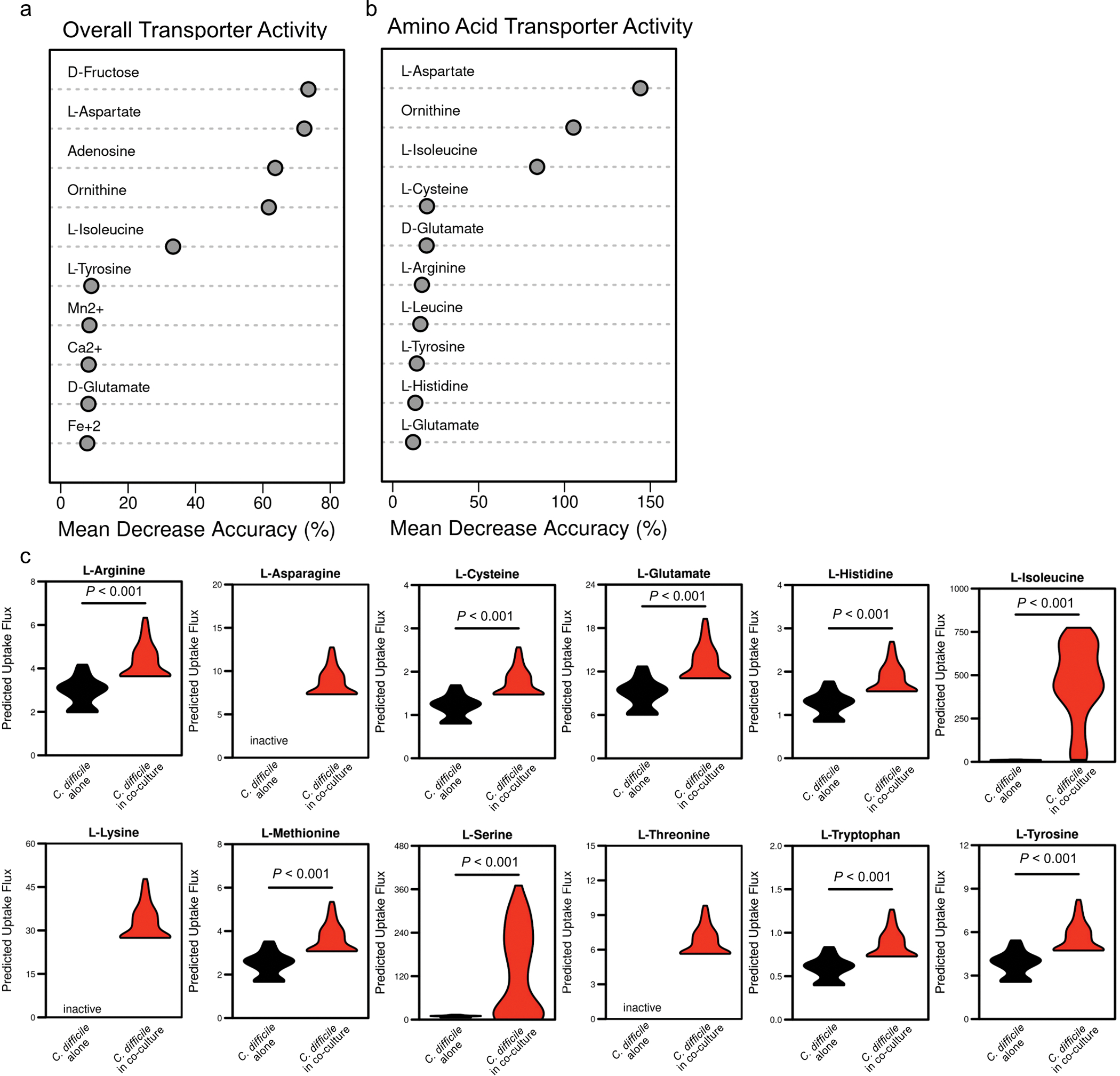
(a) AUC-Random Forest supervised machine learning results for reaction flux samples for conserved transport reactions between contexts (k = 10; OOB = 0%). (b) AUC-Random Forest supervised machine learning results for reaction flux samples for conserved amino acid transport reactions between contexts (k = 10; OOB = 0%). (c) Difference in simulated uptake of selected amino acids across context-specific models. Significance determined by two-sided Wilcoxon rank-sum test. Corrected P values in Supplementary Table 5.
