Figure 4. Macrophage-specific expression of ancestral full-length EsxM, but not its paralogs or modern, truncated EsxM, alters macrophage motility.
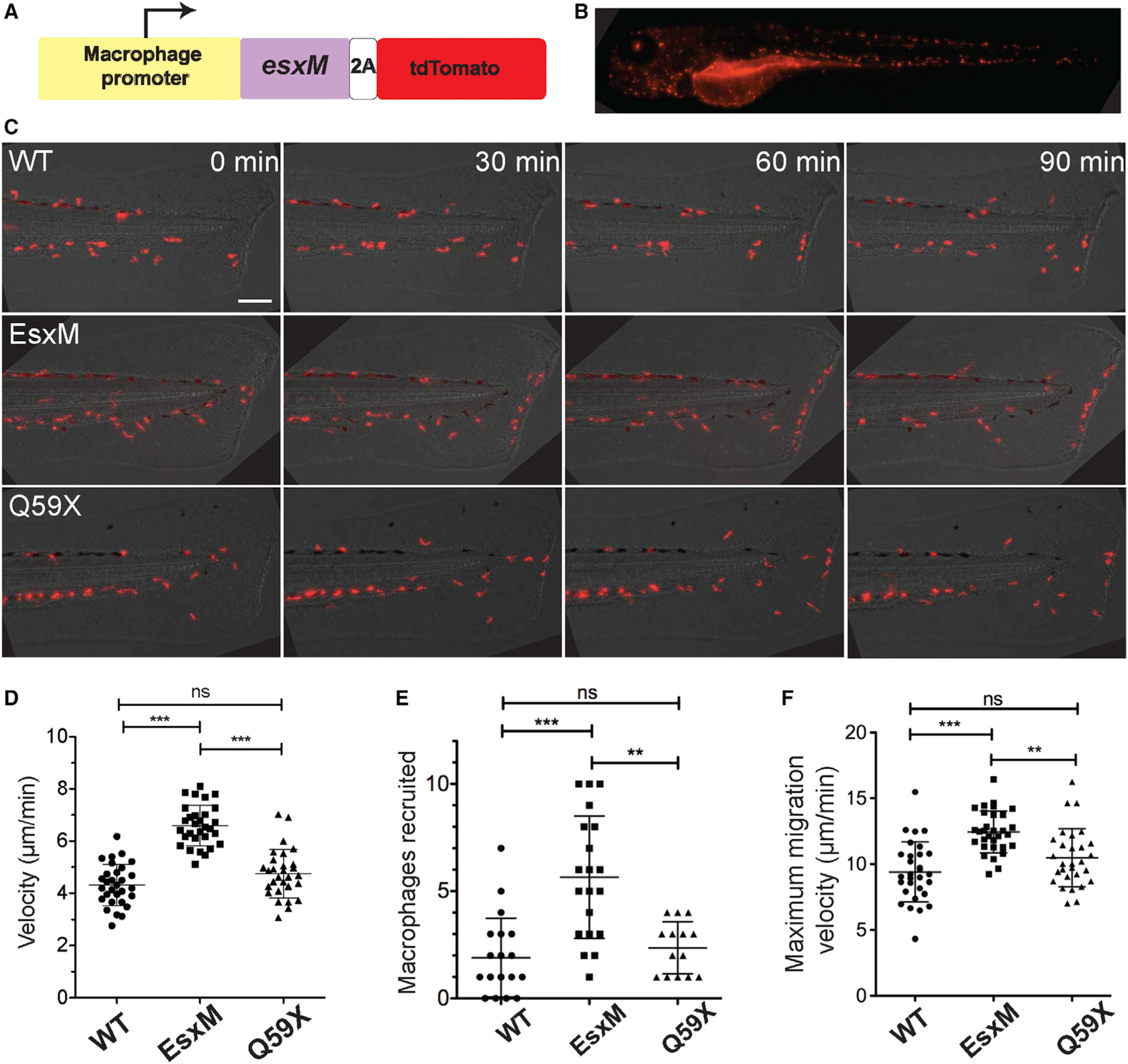
(A) Transgene design (not to scale) for macrophage-specific expression of EsxM. The macrophage-specific zebrafish promoter mfap4 drives expression of the mycobacterial protein within zebrafish macrophages. 2A peptide uncouples EsxM protein (98 AA) from tdTomato.
(B) Representative larval zebrafish expressing the mfap4:esxM-p2A-tdTomato transgene. Scale bar, 500 μm.
(C) Increased macrophage motility in the absence of infection for EsxM-expressing transgenic zebrafish lines. Scale bar, 100 μm.
(D) Migration and velocity measured in response to tail fin transection. One-way ANOVA, Dunn’s multiple comparison test. Data representative of three biological replicates.
(E) Number of macrophages recruited to tail wound 90 min post-transection. Macrophages express fluorescent TdTomato, EsxM, or truncated EsxM (Q59X) via a macrophage-specific promoter (mfap4). One-way ANOVA with Dunn’s multiple comparison test. Data representative of three biological replicates.
(F) Maximum velocity of macrophages migrating to tail wound. One-way ANOVA with Dunn’s multiple comparison test. Data representative of three biological replicates. Mean ± SD shown on all graphs. **p < 0.01, ***p < 0.001.
