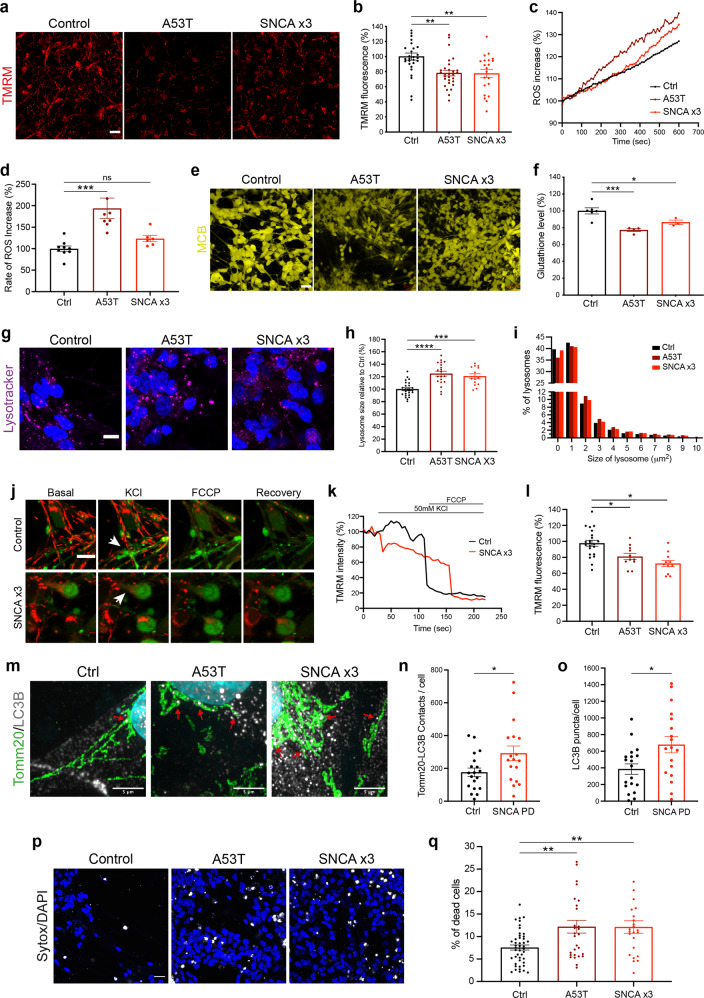
Fig. 6. Cellular dysfunction and cell death arise later in SNCA PD mDA neurons.
a Representative live-cell imaging of mitochondrial fluorescence using the lipophilic cationic dye TMRM at day 48 of differentiation. Scale bar = 10 μm. b Quantification of the normalised fluorescence intensity of TMRM (**P < 0.005, one-way ANOVA). c Trace showing the ratiometric measurement of superoxide generation using dihydroethidium (HEt) at day 48 of differentiation. d Quantification of the rate of superoxide generation based on HEt ratiometric fluorescence (ns P > 0.05, ***P < 0.0005, one-way ANOVA). e Representative live-cell imaging of endogenous glutathione using the fluorescent reporter MCB at day 48 of differentiation. Scale bar = 20 μm. f Quantification of the endogenous level of glutathione based on MCB fluorescence (*P < 0.05, ***P = 0.0009, one-way ANOVA). g Representative live-cell imaging of lysosomes and nuclear marker Hoechst 33342 at day 48 of differentiation (scale bar = 5 μm). h Quantification of the normalised average lysosomal area/size (***P = 0.0002, ****P < 0.0001, one-way ANOVA). i Histogram plot showing the percentage of total lysosomes in each set area bin (0–10 μm2). j Representative time series snapshots of TMRM (red) and Fluo-4 (green) in day 62 old neurons showing the response to KCl and FCCP. The arrow in the control cell highlights polarised mitochondria and calcium response to KCl and FCCP. Arrowhead in the SNCA x3 cell highlights KCL-induced TMRM intensity decrease. Scale bar = 10 μm. k Representative single-cell trace showing TMRM intensity in response to KCl and FCCP in control and SNCA x3 day 62 old neurons. l Quantification showing the decrease in TMRM intensity after KCl stimulation in control, A53T and SNCA x3 neurons (*P < 0.05, one-way ANOVA). m Instant Structured illumination microscopy (iSIM) images of control, A53T, and SNCA x3 day 55 neurons probed for mitochondrial marker Tomm20, and the autophagosome marker LC3B. Scale bar = 5 μm). n Quantification of the number of Tomm20-LC3B colocalizations per cell (*P < 0.05, Welch’s t test). o Quantification of the number of LC3B puncta per cell (*P < 0.05, Welch’s t test). p Live-cell images depicting dead cells in mDA neurons at >day 48 of differentiation using the fluorescent dye SYTOX green (scale bar = 20 μm). q Quantification of the percentage of dead cells (**P < 0.005, one-way ANOVA). All values are plotted as ±s.e.m. All N numbers for each experiment can be found in Supplementary Table 5.