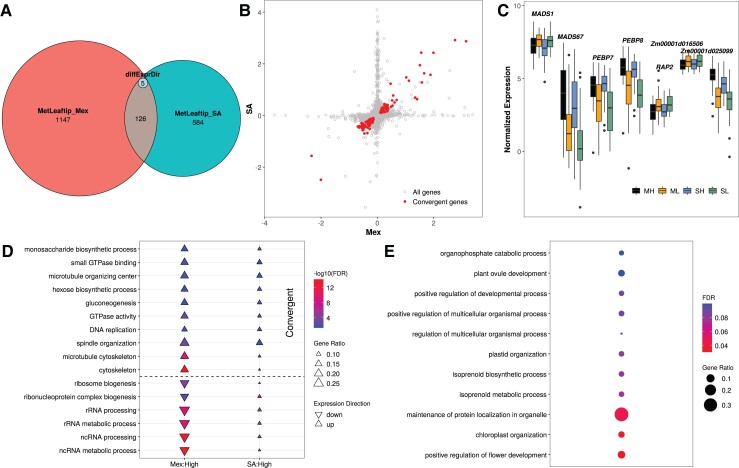
Fig. 2.
Results of gene expression analyses. (A) Numbers of differentially expressed genes between highland and lowland populations from Mexico and South America and common genes detected in both continents in the MetLeaftip tissue. The small inset in the overlapping region shows genes significant in both populations, but with opposite directions of expression change. (B) Correlation of Posterior Mean highland effects between Mexican and South American population for all genes measured for gene expression and a subset of genes showing evidence of convergent evolution (highlighted in red) in the MetLeaftip tissue. (C) Expression of flowering-related genes in the Mexican Highland (ML), Mexican Lowland (ML), South American Highland (SA), and South American Lowland (SL) populations in the MetLeaftip tissue. These flowering-related genes are identified by looking for overlapping between the convergent genes and maize flowering time candidate genes aggregated by Li et al. (2016) and Swarts et al. (2016). (D) FDR of 16 GO terms that are significant in both Mexican and South American populations across three site:tissue. The size of each triangle indicates the enrichment ratio of this GO term, defined as ratio of number of differentially expressed genes in a GO category divided by the size of the category. We tested up-regulated and down-regulated differentially expressed genes separately and triangles and upside-down triangles represent up-regulated and down-regulated GO categories, respectively. (E) GO categorical enrichments of the genes individually classified as having convergent expression evolution in MetLeaftip and MetLeafbase.