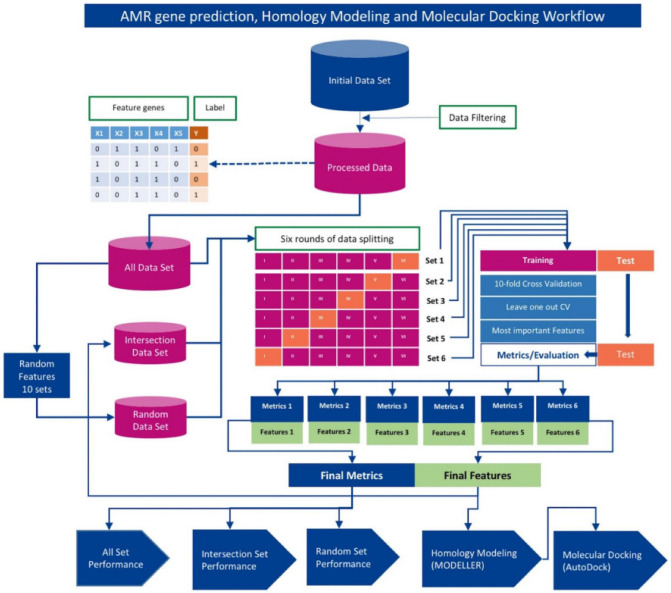
Figure 1.
Schematic diagram of AMR trait and gene prediction pipeline that integrates machine learning, homology modeling, and molecular docking. The input data matrix contained gene presence/absence information (Xi for ith gene; 1 denotes presence and 0 denotes absence) and phenotype information (Y; 1 denotes resistant and 0 denotes susceptible) for each strain (each row in the matrix). Assessment of machine learning (ML) algorithms was conducted using (i) All Set: entire gene dataset, (ii) Intersection Set: genes deemed important for discrimination and appeared consistently in all 6 rounds of cross-validation, and (iii) Random Set: randomly sampled genes (same number of genes as in Intersection set). Six-fold cross-validation was performed yielding performance of machine learning algorithms on respective data sets (i–iii). The Intersection Set genes were found to be yielding overall optimal performance and were therefore subjected to homology modeling and molecular docking analyses to assess whether protein-products of these genes form stable conformations with corresponding ligands (antibiotics) in molecular simulations.