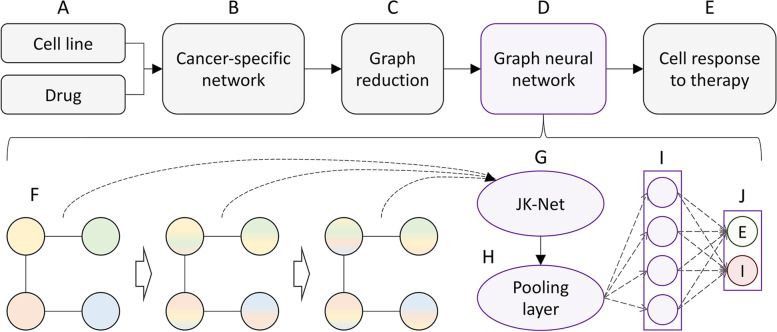
Fig. 1.
Flowchart of CancerOmicsNet. A The input consists of a cancer cell line and a drug. B The graph representation of the input data integrating a PPI network, differential gene expression, kinase inhibitor profiling, and gene-disease associations. C A graph reduction procedure guided by the topological information and the biological knowledge. D The graph neural network utilizing attention propagation mechanisms. E The predicted response of the cell line to the drug treatment. F A series of customized graph convolution blocks to generate node embeddings with information propagation steps represented by hollow arrows. The information initially carried by individual nodes is shown in different colors. G The JK-Net combining node embeddings produced by individual blocks. H A global pooling layer integrating node embeddings into the graph embedding. I A set of fully connected layers. J An output layer predicting the cell response to the drug treatment (E – effective, I – ineffective)