Figure 2.
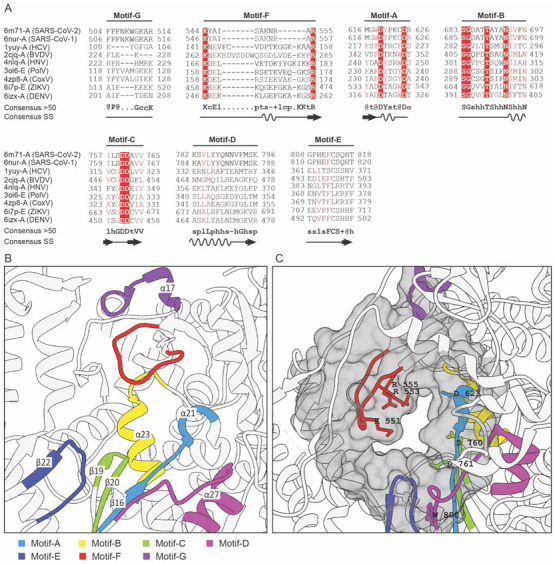
Analysis of the conserved motifs in NSP-12 and identification of druggable cavity. (A) Multiple sequences and secondary structure alignment of the homologous protein about NSP-12 of SARS-CoV-2 shows the conserved residues and secondary structure responsible for RdRp activity. Residues that have been highlighted in red are highly-conserved amino acids in the functional domain of RdRP protein. (B) Structurally and functionally conserved seven motifs A–G located at the active site of the RdRp domain of NSP-12. (C) The predicted druggable cavity at the active site of the RdRp domain of NSP-12. The predicted active residues participating in ligand interactions have been shown as a gray surface where the conserved residues are also located.
