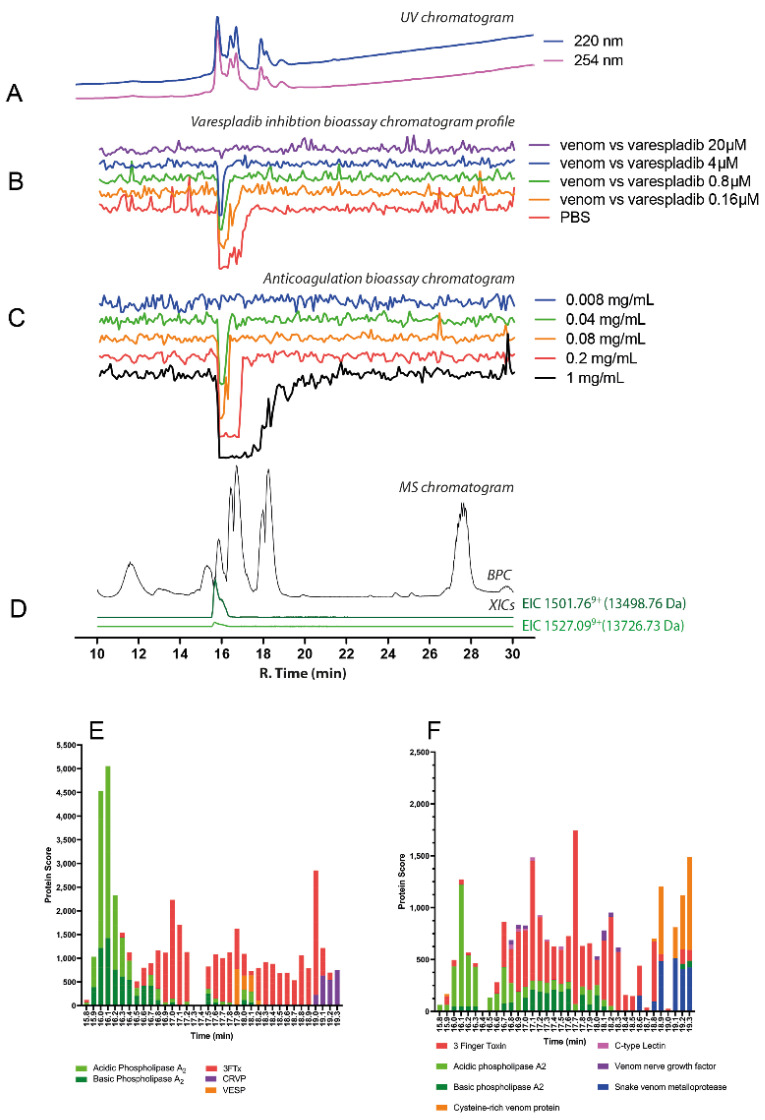
Figure 10.
Correlation of LC-UV and MS chromatogram and bioassay chromatogram of H. Haemachatus venom. (A) UV chromatogram of H. haemachatus venom (1 mg/mL, 50 μL injection volume, post-column split into 1:9 ratio. The smaller portion went to UV (220 and 254 nm recorded), and then to MS detection. Superimposed bioassay chromatograms resulting from analyses of H. haemachatus venom (0.2 mg/mL, 50 µL injection volume) in the presence of different concentrations of (B) varespladib. (C) Superimposed anticoagulation bioassay chromatograms resulting from analyses of several diluted H. haemachatus venom samples ranging from 1 mg/mL to 0.008 mg/mL (50 µL per injection). (D) Base peak chromatogram (BPC) and extracted ion currents (XIC) mass spectrometry. Bioassay chromatograms are correlated to UV and MS. Proteomic annotation of H. haemachatus venom using Mascot software against (E) the Swiss-Prot database and (F) species-specific venom gland transcriptomic-derived database. The protein score represents the probability that designated proteins, defined by elution time, are present in the sample. Note: after nanofractionation analytics, no procoagulant toxin peaks were observed (not shown in the figure). SVSPs and other mainly larger toxins can denature during the chromatographic separation thereby losing their biological activity (which we have observed in other ongoing research projects).