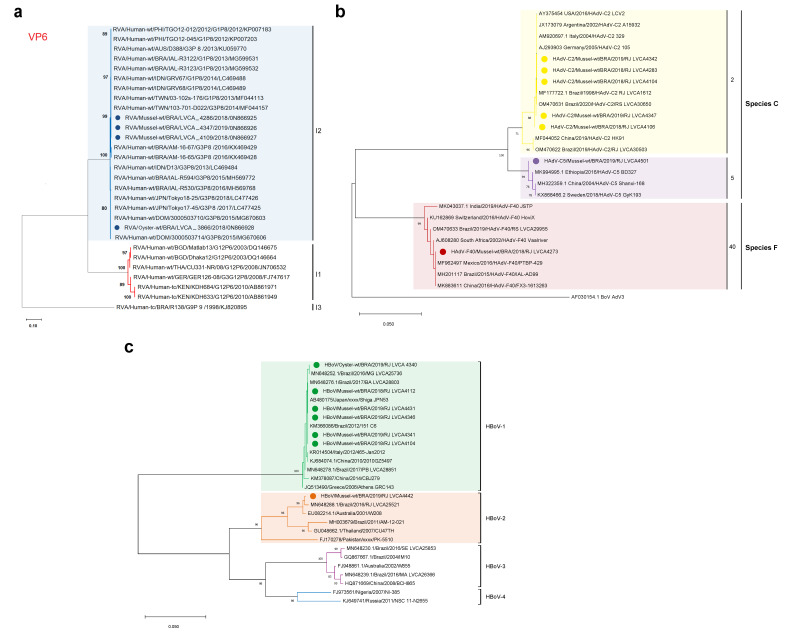
Figure 4.
Phylogenetic trees based on (a) VP6 nucleotide (nt) of RVA; on (b) the hexon gene of HAdV; and on (c) VP1 nt of HBoV. RVA strains obtained in this study are marked with a blue-filled circle. HAdV-C2, -C5 and -F40 are marked with a yellow, purple and red-filled circle, respectively. HBoV-1 and -2 are marked with a green- and orange-filled circle, respectively. Reference strains were downloaded from GenBank and labelled with their accession number, country of isolation, year, genotype and register number. Phylogenetic trees were generated in MEGA X software with bootstrap tests of 2000 replicates, based on the Tamura 3-parameter model (best fit model for all viruses). Bootstrap values of ≥70% are shown at each branch point.