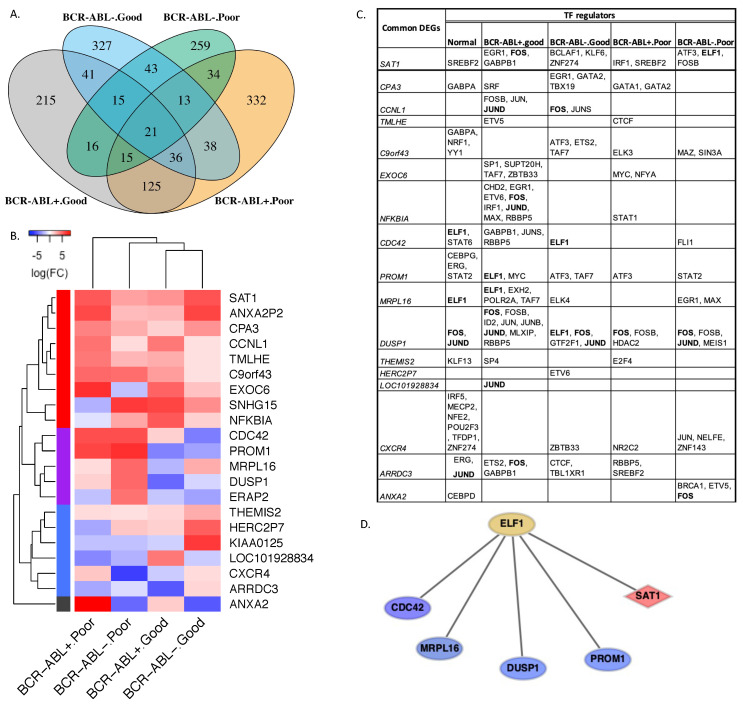
Figure 1.
Gene expression and regulation changes in different CP-CML stem cells. (A) The pairwise comparisons between normal HSCs and different stem cell groups, which were BCR-ABL+ and BCR-ABL− of good responders as well as BCR-ABL+ and BCR-ABL− of poor responders, respectively, resulted in four differentially expressed gene sets. (B) The hierarchical clusters of the stem cell groups were based on expression changes of 21 common differentially expressed genes. The cells from the patients with the same TKI response (either good or poor) were clustered together. Meanwhile, the genes formed four clusters (red, purple, blue, and grey) according to their expression alteration patterns in different cell groups as compared to normal HSCs. (C) The TF regulators of common DEGs were identified from cellular transcription networks. The bold gene names in the table represent the most frequent TF regulators of common DEGs. (D) The transcription regulon of ELF1, one of the most frequent regulators in the transcription networks, controlled four underexpressed genes, shown in blue nodes, in the cells of good responders or normal HSCs, and one overexpressed gene, shown in the red node, in the cells of poor responders.