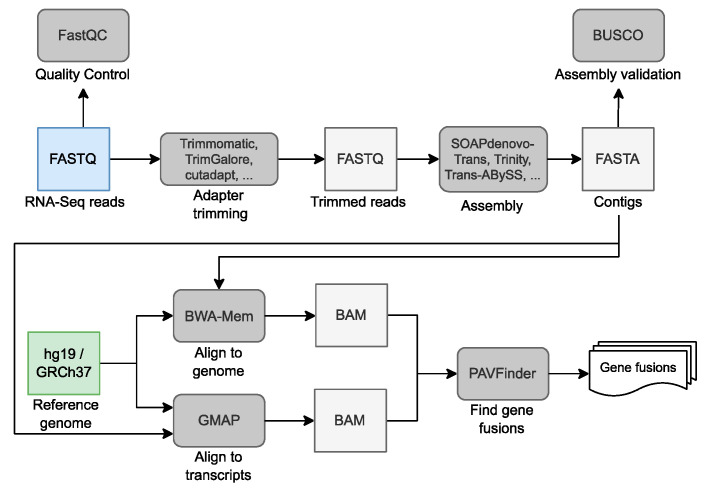
Figure 3.
Bioinformatic pipeline for gene fusion discovery through RNA-Seq data. Reads are subjected to a trimming process by means of tools such as Trimmomatic [184], cutadapt [185], or TrimGalore [186] in order to remove adapter sequences in the data. Then, trimmed reads are used for the de novo assembly process, producing assembled contigs in FASTA format. SOAPdenovo-Trans [187], Trinity [188], or Trans-ABySS [189] are examples of RNA-Seq assemblers. These contigs are aligned both to the reference genome and to the reference transcripts. Finally, resulting alignment files in BAM format were analyzed using PAVFinder [190] in order to discover transcriptomic SVs as gene fusions or tandem repeats.