Abstract
Trichoderma is known worldwide as biocontrol agents of plant diseases, producers of enzymes and antibiotics, and competitive contaminants of edible fungi. In this investigation of contaminated substrates of edible fungi from North China, 39 strains belonging to 10 Trichoderma species isolated from four kinds of edible fungi were obtained, and three novel species belonging to the Harzianum clade were isolated from the contaminated substrates of Auricularia heimuer and Pholiota adipose. They were recognized based on integrated studies of phenotypic features, culture characteristics, and molecular analyses of RNA polymerase II subunit B and translation elongation factor 1-α genes. Trichoderma auriculariae was strongly supported as a separate lineage and differed from T. vermifimicola due to its larger conidia. Trichoderma miyunense was closely related to T. ganodermatigerum but differed due to its smaller conidia and higher optimum mycelial growth temperature. As a separate lineage, T. pholiotae was distinct from T. guizhouense and T. pseudoasiaticum due to its higher optimum mycelial growth temperature and larger conidia. This study extends the understanding of Trichoderma spp. contaminating substrates of edible fungi and updates knowledge of species diversity in the group.
Keywords: Hypocreaceae, Trichoderma, phylogeny, morphology, taxonomy
1. Introduction
Trichoderma Pers. is ubiquitous in various niches and around the world. The genus contains at least eight infrageneric clades, of which the Harzianum clade is one of the largest [1]. According to our investigated statistics, the Harzianum clade consists of more than 95 accepted species, which are morphologically heterogeneous and phylogenetically complicated. They play important roles in agriculture, industry, and other fields and are employed as biocides or biofertilizers for plant growth [2,3,4], act as producers of enzymes and antibiotics, and are endophytic in plants that can resist both physiological stress and pathogen invasion [5,6].
Green mold contamination caused by Trichoderma spp. in the cultivation and various growth stages of edible fungi has been one of the biggest biological constraints in the industry since the 1980s [7], with the economic losses accounting for 10–20% of total production [8]. At present, green mold is one of the most devastating diseases in nearly all production areas of cultivated edible fungi due to its high disease incidence and serious economic loss [9,10]. Mycelia of Trichoderma spp. show stronger competitiveness than those of edible fungi, and thus they can inhibit mycelial growth or decrease the fruiting rate of edible fungi. Lots of green conidia of Trichoderma will gradually cover the contaminated substrates or fruiting bodies, and the contaminated fruiting bodies will eventually shrivel and rot.
In order to better understand the Trichoderma species contaminating substrates of edible fungi and preserve biological control resources, substrates of edible fungi contaminated by green mold in North China were investigated, and three undescribed species belonging to the Harzianum clade were found on contaminated substrates of Auricularia heimuer and Pholiota adipose. Their phylogenetic positions were determined based on sequence analyses of the combined translation elongation factor 1-alpha (tef1-α) and the second largest nuclear RNA polymerase subunit (rpb2) genes. Similarities and differences in morphological characteristics between the new species and their closely related species were investigated and compared in detail.
2. Materials and Methods
2.1. Isolates and Specimens
Specimens were separately collected from contaminated substrate of edible fungi in North China from 2020 to 2022 (Table S1), and strains were isolated following the method of a previous study [11]. The ex-type strains were deposited in the culture collection of Institute of Plant Protection, Beijing Academy of Agriculture and Forestry Sciences (JZB culture collection).
2.2. Morphology and Growth Characterization
For morphological studies, growth rates were determined on three different media: potato dextrose agar (PDA; 200 g potato, 18 g dextrose, 18 g agar, and 1 L distilled water), cornmeal dextrose agar (CMD; 40 g cornmeal, 20 g glucose, 18 g agar, and 1 L distilled water), and synthetic low nutrient agar (SNA; 1 g KH2PO4, 1 g KNO3, 0.5 g MgSO4·7H2O, 0.5 g KCl, 0.2 g glucose, 0.2 g sucrose, 18 g agar, and 1 L distilled water) at 25, 30, and 35 °C in darkness. Mycelial discs (5 mm diameter) were incubated in Petri dishes (90 mm diameter) with three replicates for each isolate. Colony diameters were measured after 3 days. The time when mycelia entirely covered the surface of the plate and the morphological characteristics of colonies, such as colony appearance, color, and spore production, were recorded [12]. For microscopic morphology, photographs were taken with an Axio Imager Z2 microscope (Carl Zeiss, Jena, Germany). Microscopic characteristics and micromorphological data were examined on the cultures grown on SNA and PDA for 7–9 days at 25 °C.
2.3. DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted from the cultures grown on PDA for 7 days using a plant genomic DNA Kit (DP305, TIANGEN Biotech, Beijing, China). Fragments of tef1-α and rpb2 were amplified with the primer pairs EF1-728F [13] and TEF1LLErev [14] and fRPB2-5f/7cr [15], respectively. Each PCR reaction consisted of 12.5 µL Premix Taq™ (TaKaRa, Dalian, China), 1.0 µL of forward primer (10 µM), 1.0 µL of reverse primer (10 µM), 1.5 µL of DNA, and 9 µL of double-sterilized water. Polymerase chain reaction (PCR) conditions followed Zhu and Zhuang [16]. The products were purified and subjected to sequencing on an ABI 3730 DNA sequencer (Applied Biosystems, Bedford, MA, USA) at SinoGenoMax company. Sequences generated from this study and those retrieved from GenBank are listed in Table 1.
Table 1.
Materials including strain numbers and GenBank accessions of sequences used for phylogenetic analyses.
| Species | Voucher | GenBank Accession Number | |
|---|---|---|---|
| rpb2 | tef1-α | ||
| T. achlamydosporum | YMF 1.6177 | MT052180 | MT070156 |
| T. afarasin | CBS 130755 | – | AF348093 |
| T. afarasin | DIS 314F | FJ442778 | FJ463400 |
| T. afroharzianum | CBS 124620 ET | FJ442691 | FJ463301 |
| T. afroharzianum | GJS 04-193 | FJ442709 | FJ463298 |
| T. aggregatum | HMAS 248863 | KY688001 | KY688062 |
| T. aggregatum | HMAS 248864 | KY688002 | KY688063 |
| T. aggressivum | CBS 100525 | AF545541 | AF348095 |
| T. aggressivum | DAOM 222156 ET | FJ442752 | AF348098 |
| T. alni | CBS 120633 ET | EU498349 | EU498312 |
| T. alpinum | HMAS 248821 T | KY687958 | KY688012 |
| T. amazonicum | IB95 | HM142368 | HM142377 |
| T. amazonicum | CBS126898 ET | HM142367 | HM142376 |
| T. anaharzianum | YMF 1.00241 | MH262577 | MH236493 |
| T. anaharzianum | YMF 1.00383 T | MH158995 | MH183182 |
| T. asiaticum | YMF 1.00168 | MH262575 | MH236492 |
| T. asiaticum | YMF 1.00352 T | MH158994 | MH183183 |
| T. atrobrunneum | GJS90-254 | FJ442735 | AF443943 |
| T. atrobrunneum | GJS 05-101 | FJ442745 | FJ463392 |
| T. atrogelatinosum | CBS 237.63 ET | KJ842201 | – |
| T. atroviride | CBS 119499 | FJ860518 | FJ860611 |
| T. auriculariae | JZBQT1Z7 T | ON649949 | ON649896 |
| T. auriculariae | JZBQT1Z8 | ON649950 | ON649897 |
| T. auriculariae | JZBQT1Z9 | ON649951 | ON649898 |
| T. austroindianum | BAFC 3583 | – | MH352421 |
| T. azevedoi | CEN1422 | MK696821 | MK696660 |
| T. bannaense | HMAS 248840 T | KY687979 | KY688037 |
| T. bannaense | HMAS 248865 | KY688003 | KY688038 |
| T. botryosum | COAD 2422 | MK044212 | MK044119 |
| T. botryosum | COAD 2401 | MK044181 | MK044088 |
| T. breve | HMAS 248844 T | KY687983 | KY688045 |
| T. breve | HMAS 248845 | KY687984 | KY688046 |
| T. brunneoviride | CBS 121130 | EU498357 | EU498316 |
| T. brunneoviride | CBS 120928 | EU498358 | EU498318 |
| T. caeruloviride | COAD 2416 | MK044201 | MK044108 |
| T. caeruloviride | COAD 2415 | MK044202 | MK044109 |
| T. camerunense | GJS 99-230 | – | AF348107 |
| T. catoptron | GJS 02-76 T | AY391900 | AY737726 |
| T. ceraceum | GJS 95-159 | AF545508 | AY937437 |
| T. ceratophylletum | YMF 1.04621 T | MK327580 | MK327579 |
| T. cerinum | DAOM 230012 | KJ842184 | KJ871242 |
| T. christiani | CBS 132572 ET | KJ665244 | KJ665439 |
| T. cinnamomeum | GJS 96-128 | AY391916 | AY391977 |
| T. cinnamomeum | GJS 97-233 | AY391919 | AY391978 |
| T. compactum | CBS 121218 | KF134789 | KF134798 |
| T. concentricum | HMAS 248833 | KY687971 | KY688027 |
| T. confertum | HMAS 248893 | MF371203 | MF371218 |
| T. confertum | HMAS 248896 | MF371205 | MF371220 |
| T. corneum | GJS 97-82 ET | KJ665252 | KJ665455 |
| T. dacrymycellum | WU29044 | FJ860533 | FJ860633 |
| T. endophyticum | CBS 130753 | FJ442722 | FJ463326 |
| T. endophyticum | CBS 130733 | FJ442690 | FJ463330 |
| T. epimyces | CBS120534 ET | EU498360 | EU498320 |
| T. ganodermatigerum | CCMJ5245 T | ON567189 | ON567195 |
| T. ganodermatigerum | CCMJ5246 | ON567190 | ON567196 |
| T. ganodermatigerum | CCMJ5247 | ON567191 | ON567197 |
| T. globoides | HMAS 248747 | KX026963 | KX026955 |
| T. guizhouense | HGUP0038 T | JQ901400 | JN215484 |
| T. guizhouense | S278 | KF134791 | KF134799 |
| T. guizhouense | DAOM 231435 | – | EF191321 |
| T. harzianum | CBS 226-95 | AF545549 | AF348101 |
| T. harzianum | GJS 05 107 | FJ442708 | FJ463329 |
| T. hausknechtii | CBS 133493 | KJ665276 | KJ665515 |
| T. helicolixii | CBS 133499 | KJ665278 | KJ665517 |
| T. hengshanicum | HMAS 248852 T | KY687991 | KY688054 |
| T. hirsutum | HMAS 248834 T | KY687972 | KY688029 |
| T. hortense | BMCC LU994 | – | KJ871185 |
| T. ingratum | HMAS 248822 | KY687973 | KY688018 |
| T. inhamatum | CBS 273-78 | FJ442725 | AF348099 |
| T. italicum | CBS 132567 | KJ665282 | KJ665525 |
| T. koreanum | SFC20131005-S066 | MH025988 | MH025979 |
| T. lentiforme | DIS 253B | FJ442756 | FJ851875 |
| T. lentiforme | DIS 94D | FJ442749 | FJ463379 |
| T. lentinulae | HMAS 248256 | MN605867 | MN605878 |
| T. lentinulae | CGMCC 3.19848 | MN605868 | MN605879 |
| T. liberatum | HMAS 248831 T | KY687969 | KY688025 |
| T. linzhiense | HMAS 248846 T | KY687985 | KY688047 |
| T. lixii | CBS 110080 | KJ665290 | FJ716622 |
| T. longifialidicum | LESF 552 | KT278955 | KT279020 |
| T. miyunense | JZBQF5 | ON649968 | ON649915 |
| T. miyunense | JZBQF7 T | ON649969 | ON649916 |
| T. miyunense | JZBQF9 | ON649970 | ON649917 |
| T. neotropicale | LA11 ET | – | HQ022771 |
| T. paratroviride | S385 | KJ665321 | KJ665627 |
| T. parepimyces | CBS 122769 ET | FJ860562 | FJ860664 |
| T. peberdyi | CEN1426 | MK696825 | MK696664 |
| T. peruvianum | CP15-2 | MW480153 | MW480145 |
| T. peruvianum | CP15-9 | MW480154 | MW480146 |
| T. perviride | HMAS 273786 | KX026962 | KX026954 |
| T. phayaoense | SDBR-CMU349 | MW002074 | MW002073 |
| T. pholiotae | JZBQH11 | ON649971 | ON649918 |
| T. pholiotae | JZBQH12 T | ON649972 | ON649919 |
| T. pholiotae | JZBQH13 | ON649973 | ON649920 |
| T. pinicola | KACC 48486 ET | MH025993 | MH025981 |
| T. pinicola | SFC20130926-S014 | MH025991 | MH025978 |
| T. pleuroti | CBS 124387 ET | HM142372 | HM142382 |
| T. pleuroticola | CBS 124383 ET | HM142371 | HM142381 |
| T. pleuroticola | TRS70 ET | KP009172 | KP008951 |
| T. pollinicola | LC11682 = LF1542 ET | MF939604 | MF939619 |
| T. pollinicola | LC11686 = LF2050 | MF939605 | MF939620 |
| T. polypori | HMAS 248855 T | KY687994 | KY688058 |
| T. priscilae | CBS 131487 ET | KJ665333 | KJ665691 |
| T. propepolypori | YMF 1.06224 T | MT052181 | MT070158 |
| T. propepolypori | YMF 1.06199 | MT052182 | MT070157 |
| T. pseudoasiaticum | YMF 1.06200 T | MT052183 | MT070155 |
| T. pseudodensum | HMAS 248828 T | KY687967 | KY688023 |
| T. pseudogelatinosum | CNUN309 ET | HM920173 | HM920202 |
| T. pseudopyramidale | COAD 2419 | MK044206 | MK044113 |
| T. pseudopyramidale | COAD 2506 | MK044207 | MK044114 |
| T. purpureum | HMAS 273787 T | KX026961 | KX026953 |
| T. pyramidale | CBS 135574 ET | KJ665334 | KJ665699 |
| T. rifaii | CBS 130746 | – | FJ463324 |
| T. rifaii | DIS 337F | FJ442720 | FJ463321 |
| T. rufobrunneum | HMAS 266614 T | KF730010 | KF729989 |
| T. rugulosum | SFC20180301-001 T | MH025986 | MH025984 |
| T. rugulosum | SFC20180301-002 | MH025987 | MH025985 |
| T. simile | YMF 1.06201 T | MT052184 | MT070154 |
| T. simile | YMF 1.06202 | MT052185 | MT070153 |
| T. simmonsii | CBS 130431 | FJ442757 | AF443935 |
| T. simmonsii | S7 | KJ665337 | KJ665719 |
| T. simplex | HMAS 248842 T | KY687981 | KY688041 |
| T. solum | HMAS 248848 T | KY687987 | KY688050 |
| T. stramineum | GJS 02-84 | AY391945 | AY391999 |
| T. subalni | HMAS 275683 | MH612371 | MH612377 |
| T. subalni | HMAS 275684 | MH612370 | MH612376 |
| T. syagri | BAFC 4357 | – | MG822711 |
| T. tawa | CBS 114233 ET | AY391956 | FJ463313 |
| T. tawa | DAOM 232841 | KJ842187 | EU279972 |
| T. tenue | HMAS 273785 ET | KX026960 | KX026952 |
| T. tomentosum | DAOM 178713a | AF545557 | AY750882 |
| T. velutinum | CPK 298 | KF134794 | KJ665769 |
| T. velutinum | DAOM 230013 ET | JN133569 | AY937415 |
| T. vermifimicola | CGMCC 3.19850 | MN605870 | MN605881 |
| T. vermifimicola | HMAS 248255 | MN605871 | MN605882 |
| T. xixiacum | HMAS 248253 T | MN605874 | MN605885 |
| T. xixiacum | CGMCC 3.19698 | MN605875 | MN605886 |
| T. zayuense | HMAS 248835 T | KY687974 | KY688031 |
| T. zelobreve | HMAS 248254 T | MN605872 | MN605883 |
| T. zelobreve | CGMCC 3.19696 | MN605873 | MN605884 |
| T. zeloharzianum | YMF 1.00268 | MH158996 | MH183181 |
Numbers in bold indicate newly submitted sequences in this study. T: type strains. ET: ex-type strains.
2.4. Phylogenetic Analyses
Sequences for all isolates generated in this study were blasted against the NCBIs GenBank nucleotide datasets (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and MIST (http://mmit.china-cctc.org/index.php) [17] to obtain an initial identification. To identify the phylogenetic positions of Trichoderma species isolated from contaminated substrates of edible fungi, rpb2 and tef1-α sequences of all Trichoderma species belonging to the Harzianum clade were combined for the analyses, with T. atroviride and T. paratroviride selected as outgroup taxa. Their sequences of type or ex-type strains based on previous publications were downloaded from NCBI database and assembled using BioEdit 7.0.5.3 [18]. Alignment was generated and converted to nexus files with Clustal X 1.83 [19].
Maximum parsimony (MP) analysis was performed with PAUP 4.0b10. Starting trees were obtained via random stepwise addition with 1000 replicates and subsequent branch-swapping algorithm using tree bisection–reconnection (TBR) [20]. Analyses were performed with all characters treated as unordered and unweighted, and gaps treated as missing data. MaxTrees was set to 1000, and branches collapsed when maximum branch length was zero. Maximum parsimony bootstrap proportion (MPBP) was calculated to test topological confidence of the resulting trees.
Bayesian inference (BI) trees were calculated using MrBayes v. 3.1.2 [21]. The best-fit nucleotide substitution model GTR+I+G was selected using MrModeltest 2.3 [22]. Four chains were run from random trees for 6,000,000 generations and sampled every 100 generations. The first 25% of trees were discarded as the burn-in phase of the analyses, and Bayesian inference posterior probability (BIPP) was determined from the remaining trees. Trees were visualized in FigTree v1.4.3 [23].
3. Results
3.1. Phylogenetic Analyses
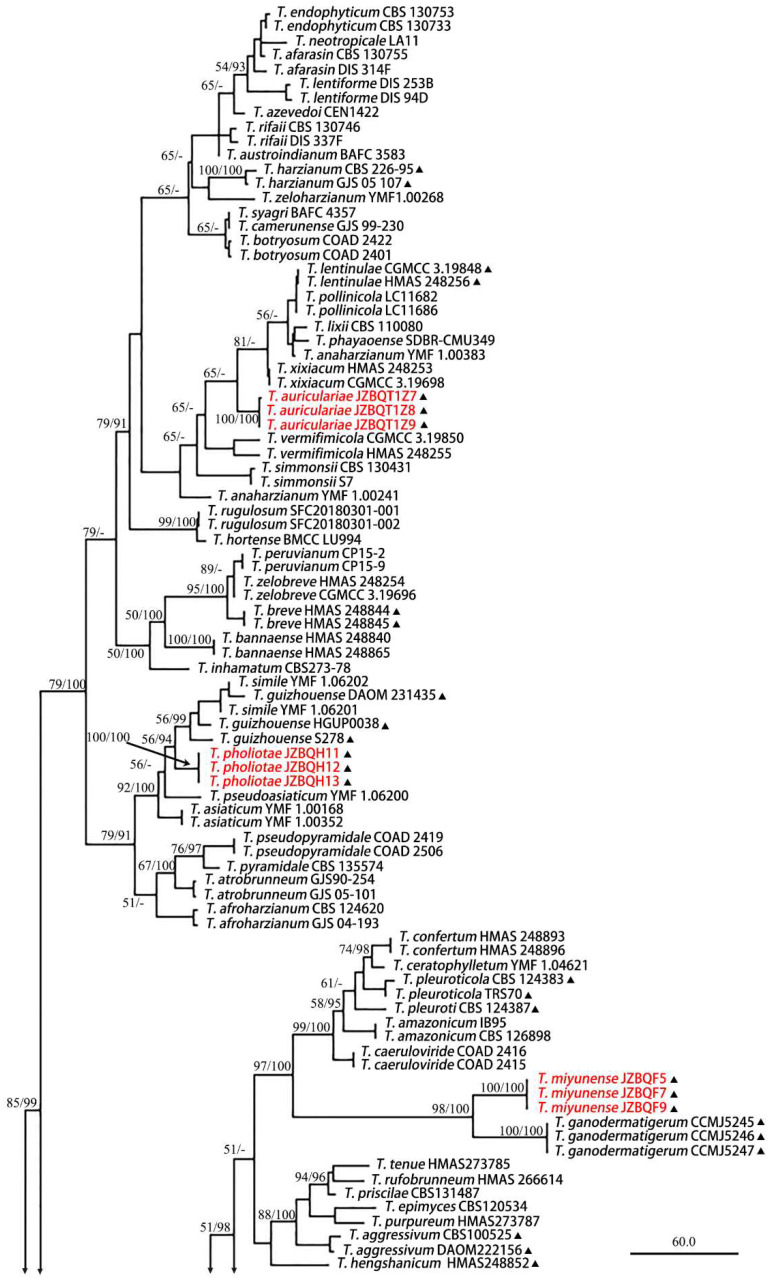
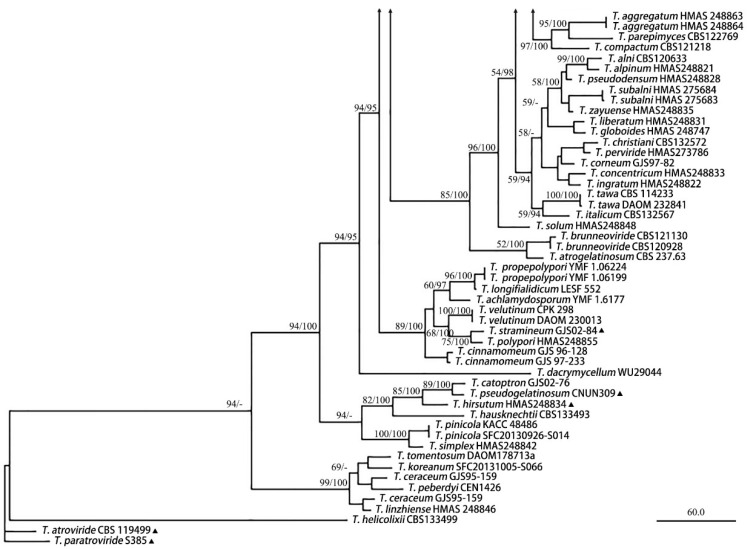
The partition homogeneity test of rpb2 and tef1-α sequences indicated that the individual partitions were generally congruent (p = 0.01). The combined rpb2 and tef1-α dataset was subsequently used for phylogenetic analysis to determine the positions of the new species. In MP analysis, the dataset contained 140 taxa and 2307 characters, of which 1468 characters were constant, 150 variable characters were parsimony uninformative, and 689 were parsimony informative. Five most parsimonious trees with the same topology were generated, and one of them is shown in Figure 1 (tree length = 3091, CI = 0.3999, HI = 0.6001, RC = 0.3039, and RI = 0.7600). The BI tree topology was generally the same as that of the MP tree.
Figure 1.
Maximum parsimony phylogram of the investigated Trichoderma species inferred from the combined sequences of rpb2 and tef1-α. MPBP above 50% (left) and BIPP above 90% (right) are indicated at the nodes. New species proposed are indicated in red font. Trichoderma species isolated from substrate or fruiting bodies of edible fungi are marked with ▲.
A total of 140 sequences representing 95 Trichoderma species, including our three new species, were used for constructing the phylogenetic tree, and T. atroviride and T. paratroviride were used as outgroups. Results showed that all the investigated Trichoderma species formed a strongly supported group (MPBP/BIPP = 100%/100%), which was generally congruent with the previous studies [24].
In the phylogenetic tree (Figure 1), T. auriculariae, T. miyunense, and T. pholiotae were newly added to the T. harzianum clade. Trichoderma auriculariae was distributed as a separate terminal branch (MPBP/BIPP = 100%/100%) among T. vermifimicola and T. xixiacum. Trichoderma miyunense was a sister of T. ganodermatigerum (MPBP/BIPP = 98%/100%). Trichoderma pholiotae formed a linage with T. asiaticum, T. guizhouense, T. pseudoasiaticum, and T. simile with high support value (MPBP/BIPP = 92%/100%), and our three strains of T. pholiotae were distributed as a highly supported separate terminal branch (MPBP/BIPP = 100%/100%) among T. pseudoasiaticum and T. guizhouense.
3.2. Taxonomy
Trichoderma auriculariae Z. J. Cao and W.T. Qin, sp. nov.
MycoBank MB845141 (Figure 2).
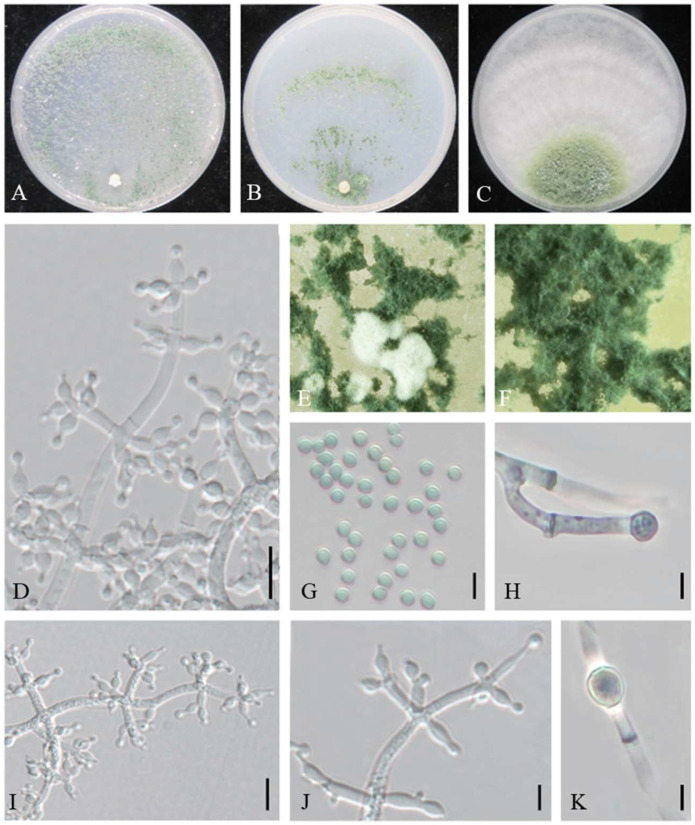
Figure 2.
Trichoderma auriculariae (JZBQT1Z7). Cultures at 25 °C after 7 days on (A) CMD, (B) SNA, and (C) PDA; (D,I,J) conidiophores and phialides; (E,F) conidiation pustules on CMD after 7 days; (G) conidia; (H,K) chlamydospores. Scale bars: (D,I) = 10 µm, (G,H,J,K) = 5 µm.
Etymology: The specific epithet refers to the host from which the fungus was isolated.
Typification: China, Beijing, Tongzhou, from the contaminated substrates of Auricularia heimuer, 26 August 2021, W.T. Qin, Z.J. Cao, L. Gao, J. Li (ex-type strain JZBQT1Z7).
DNA barcodes: ITS = ON653396, rpb2 = ON649949, tef1-α = ON649896.
On CMD after 72 h, colony radius 65–66 mm at 25 °C, 69–70 mm at 30 °C, and 8–10 mm at 35 °C. Colony hyaline and radial, not zonate. Aerial hyphae rare in colony center. A large number of white pustules formed after 2 days. Conidiation formed on aerial hyphae and in pustules, abundant, spreaded throughout the colony, then gradually turned green. No diffusing pigment noted.
On PDA after 72 h, colony radius 47–49 mm at 25 °C, 66–68 mm 30 °C, and 5–7 mm at 35 °C. Colony regularly circular, distinctly zonate. Aerial mycelium dense and radial, forming a dense, zonate, floccose mat. Conidial production noted after 2 days, starting around the original inoculum, effuse in aerial hyphae, more abundant along the original inoculum. No diffusing pigment noted, odor fruity.
On SNA after 72 h, colony radius 47–49 mm at 25 °C, 51–55 mm at 30 °C, and 5–7 mm at 35 °C. Colony hyaline, mycelium loose. Conidial production noted after 2 days, starting around the inoculum, effuse in the aerial hyphae, forming a few inconspicuous rings. Small pustules formed around the inoculum, first white, turning green after 3 days, with hairs protruding beyond the surface. No diffusing pigment.
Conidiophores pyramidal, with opposing branches borne on a conspicuously broad spindle, less solitary. The main axis and branches terminating in 3–5 cruciate to nearly verticillate disposed phialides. Hyphal septa clearly visible. Phialides ampulliform, sometime lageniform, 4.6–9.9 × (2.2–) 2.7–3.8 µm, l/w 1.4–3.5 (–4.4), 1.4–2.7 µm wide at the base (n = 50). Conidia green, globose or subglobose, sometimes ellipsoidal, smooth, 2.7–3.8 × 2.3–3.1 µm, l/w 1.0–1.3 (n = 50). Chlamydospores common, intercalary or terminal, variable in shape, ellipsoid, globose or oblong, 4.6–7.5 × 3.8–6.3 µm (n = 20).
Additional strains examined: China, Beijing, Tongzhou, from the contaminated substrates of A. heimuer, 26 August 2021, W.T. Qin, Y. Liu, S.X. Wang, JZBQT1Z8; ibid., JZBQT1Z9.
Notes: Phylogenetically, T. auriculariae formed a separate group (MPBP/BIPP = 100%/100%) in the Harzianum clade among T. vermifimicola and T. xixiacum. The tef1-α sequences between T. auriculariae and T. vermifimicola were very similar, but they shared 28 bp divergent among 1117 bp for rpb2 sequences (97.49%). Phylogenetically, T. auriculariae shared a common ancestor with T. xixiacum, T. vermifimicola, and T. simmonsi. Trichoderma auriculariae shared typical characteristics of the Harzianum clade in pyramidal conidiophores comprising a long main axis, and 3–5 phialides in whorls arose at the tips of the branches. However T. auriculariae had longer phialides and grew much slower at 35 °C on PDA than T. simmonsi [5.2–6.5 mm, 25–55 mm] [25] and had larger conidia than that of T. vermifimicola [2.3–2.6 × 2.0–2.4 µm] and T. xixiacum [2.3–2.7 × 2.0–2.6] [24]. Meanwhile, chlamydospores were common in T. auriculariae (Table S1).
Trichoderma miyunense Z. J. Cao and W.T. Qin, sp. nov.
MycoBank MB845142 (Figure 3).
Figure 3.
Trichoderma miyunense (JZBQF9). Cultures at 25 °C after 7 days on (A) CMD, (B) SNA, and (C) PDA; (D,F,H) conidiophores and phialides; (E,G) conidiation pustules on SNA after 7 days; (I) chlamydospores. Scale bars: (D,H) = 10 µm, (F,I) = 5 µm.
Etymology: The specific epithet refers to the type locality.
Typification: China, Beijing, Miyun, from the contaminated substrates of Auricularia heimuer, 9 September 2020, Y. Liu, W.T. Qin, S. Song (ex-type strain JZBQF9).
DNA barcodes: ITS = ON653404, rpb2 = ON649970, tef1-α = ON649917.
On CMD after 72 h, colony radius 51–52 mm at 25 °C and 65–66 mm at 30 °C. No growth at 35 °C. Colony hyaline, weak, regularly circular, distinctly zonate. Conidiation first formed in white pustules on aerial hyphae, turned green after a few days. No diffusing pigment noted, odor slightly fruity.
On PDA after 72 h, colony radius 42–43 mm at 25 °C and 51–54 mm at 30 °C. No growth at 35 °C. Mycelium white, aerial along the edge, irregularly circular, less with sporulation. No diffusing pigment noted, odor slightly fruity.
On SNA after 72 h, colony radius 30–33 mm at 25 °C and 25–29 mm at 30 °C. No growth at 35 °C. Mycelium hyaline and smooth, dark green to light green pustules, irregular in shape, relatively abundant in the zonation regions, with the formation of 2–3 concentric rings. Aerial hyphae short and inconspicuous. No diffusing pigment, no distinct odor.
Conidiophores pyramidal, with a relatively obvious main axis, multiple branches unpaired, with the longest branches near the base of the main axis. Branches perpendicular to the main axis or at acute angles with the main axis, with septa conspicuous and producing barrel-shaped or cylindrical metulae. Phialides densely disposed at the terminal of branches, often formed in whorls of 2–4, variable in shape and size, ampulliform to lageniform, (5.2–) 5.6–9.7 (–10.3) × 1.9–3.2 (–3.7) µm, l/w 1.9–4.4, 1.0–2.1 (–2.6) wide at the base (n = 80). Conidia green, smooth, ellipsoid, sometimes globose to subglobose, 2.2–3.4 × (1.8–) 2–2.9 µm, l/w 1–1.3 (–1.4) (n = 80). Chlamydospores unobserved.
Additional strains examined: China, Beijing, Miyun, from the contaminated substrates of Auricularia heimuer, 9 September 2020, W.T. Qin, Y. Liu, S. Song, JZBQF5; ibid., JZBQF7.
Notes: Phylogenetically, T. miyunense formed a sister group with T. ganodermatigerum (Figure 1). They shared 36 bp divergent among 1132 bp for rpb2 sequences (96.82%) and 35 bp divergent among 1102 bp for tef1-α sequences (96.82%). Morphologically, compared to T. miyunense, T. ceratophylletum possessed shorter phialides (4.1–8.4 µm) and lesser l/w of phialides [(1.0–) 1.2–2.8 (–3.2) µm] [26], while T. ganodermatigerum had larger conidia [(3.4–) 3.6–4.8 (–5.3) × (2.9–) 3.2–4.3 (–4.6)], and the optimum temperature was 25 °C [27]. T. miyunense was distinctly different from T. caeruloviride, which possessed abundant chlamydospores on CMD after 4 days with no concentric rings present [28]. In contrast, T. confertum had slightly larger phialides [8.3–12.5 × 2.5–4.2 µm] [29], T. amazonicum had distinctly wider phialides [3.3–3.5 µm] and chlamydospore-like structures in the clusters, and T. pleuroticola featured diffuse brown pigment and yellow crystals on PDA [30] (Table S2).
Trichoderma pholiotae Z.J. Cao & W.T. Qin, sp. nov.
MycoBank MB845143 (Figure 4).
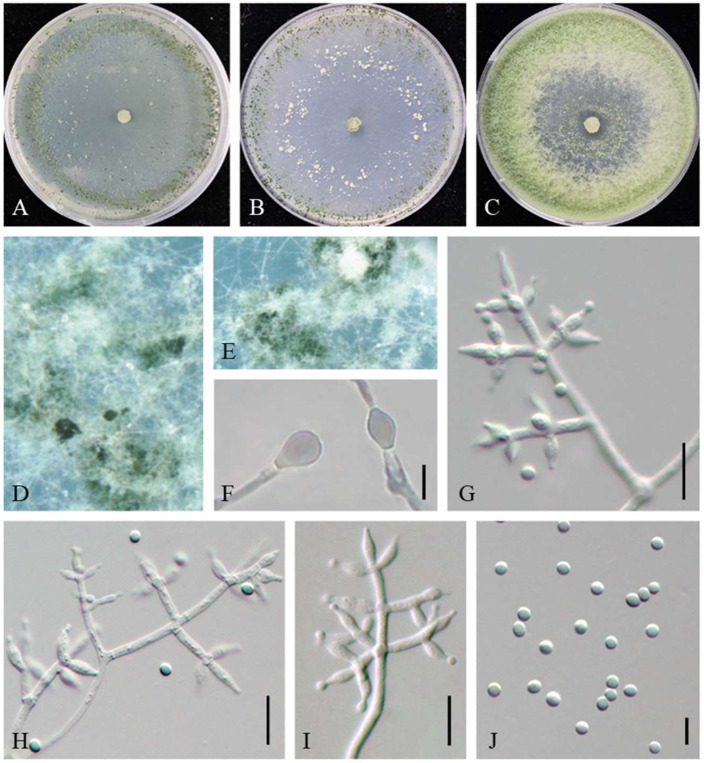
Figure 4.
Trichoderma pholiotae (JZBQH12). Cultures at 25 °C after 7 days on (A) CMD, (B) SNA, and (C) PDA; (D,E) conidiation pustules on SNA after 7 days; (F) chlamydospores; (G–I) conidiophores and phialides; (J) conidia. Scale bars: (F,J) = 5 µm, (G–I) = 10 µm.
Etymology: The specific epithet refers to the host from which the fungus was isolated.
Typification: China, Beijing, Haidian, from the contaminated substrates of Pholiota adipose, 25 September 2020, W.T. Qin, Z.J. Cao, L. Gao, J. Li (ex-type strain JZBQH12).
DNA barcodes: ITS = ON653405, rpb2 = ON649972, tef1-α = ON649919.
On CMD after 72 h, colony radius 71–72 mm at 25 °C, 73–74 mm at 30 °C, and 13–18 mm at 35 °C. Colonies hyaline, fan-shaped, tending to aggregate toward the distal parts of the colony. Aerial hyphae loose, sparse, radial. Conidiation effuse in aerial hyphae or in loosely disposed pustules. Pustules minute, irregular in shape, relatively abundant in the zonation regions, formed concentric rings around the outer ring, white at first, then gradually green. No diffusing pigment noted, odor slightly fruity.
On PDA after 72 h, colony radius 67–68 mm at 25 °C, 70–72 mm at 30 °C, and 8–10 mm at 35 °C. Colonies white in the center, with the zone around the central part of the colony forming a distinct circular and green part. Aerial hyphae distinctly radial, abundant, dense, floccose to cottony. Light diffusing yellow pigment, odor slightly fruity.
On SNA after 72 h, colony radius 49–50 mm at 25 °C, 54–55 mm at 30 °C, and 8–10 mm at 35 °C. Colonies translucent and round-like. Aerial hyphae short, radial distribution. Pustules abundant, irregular in shape, from white to green, with the formation of concentric rings. No diffusing pigment noted.
Conidiophores typically pyramidal with opposing branches, formed densely intricate reticulum, with one terminal whorl of generally 3–4 phialides and mostly paired side branches, less frequently solitary. Branches mostly perpendicular to the main axis with septa conspicuous. Phialides varied, borne in regular levels around the axis, some regular ampulliform or lageniform and others apex and inequilateral to curved, (4.1–) 4.9–10.9 (–11.6) × 2.4–4.2 (–5.0) µm, l/w 1.4–3.4 (–3.9), (1.3–) 1.4–3.1 (–3.4) µm wide at the base (n = 100). Conidia elliptic to subspheroidal, less globose, green, smooth, 2.6–3.8 (–4.2) × 2.4–3.3 (–3.5) µm, l/w 1–1.3 (n = 80). Chlamydospores common, intercalary or terminal, ellipsoid, globose, 5.0–7.4 (8.3) × (3.9–) 4.9–7.0 µm (n = 25).
Additional strains examined: China, Beijing, Haidian, from the contaminated substrates of Pholiota adipose, 25 September 2020, W.T. Qin, Z.J. Cao, L. Gao, J. Li, JZBQH11; ibid., JZBQH13.
Notes: Phylogenetically, T. pholiotae formed a linage with T. asiaticum, T. guizhouense, T. pseudoasiaticum, and T. simile with high support value (MPBP/BIPP = 92%/100%), and our three strains of T. pholiotae were distributed as a highly supported separate terminal branch (MPBP/BIPP = 100%/100%) among T. pseudoasiaticum and T. guizhouense in the Harzianum clade. However, compared to T. pholiotae, T. guizhouense possessed thinner phialides [2.0–3.0 µm] and globose conidia [31]. T. simile had distinct lower optimum growth temperature (25 °C) in the three media, and T. asiaticum had shorter phialides [(3.0–) 4.0–6.0 (–7.0) µm] [12]. In addition, T. pholiotae and T. pseudoasiaticum could be distinguished by the branching pattern, with T. pholiotae being pyramidal and T. pseudoasiaticum being verticillium-like (Table S3).
4. Discussion
During exploration of contaminated substrates of edible fungi in North China, 39 strains representing 10 Trichoderma species were isolated from four kinds of edible fungi and examined, and three new species were recognized based on integrated studies of phenotypic and molecular data (Table S1). To explore their taxonomic positions, a phylogenetic tree containing all species of the Harzianum clade was constructed based on analyses of the combined sequences of rpb2 and tef1-α. The three new species were well located in the Harzianum clade with separate terminal branches and were clearly distinguishable from any of the existing species. The results of this study have a number of practical implications to identify and diagnose Trichoderma species contaminating edible fungi. This work provides useful information on the epidemiological and geographical distribution of Trichoderma, which will help in the development of targeted interventions aimed at comprehensive management and control of green mold contamination of edible fungi.
With further study of Trichoderma classification, researchers have reached a consensus that accurate identification of Trichoderma species cannot depend only on the morphological identification as sometimes there is high ambiguity in the morphological features of Trichoderma spp. [32,33]. Trichoderma spp. isolated from the fruiting bodies or substrates of edible fungi is usually anamorph with high morphological similarity with many species, which is not conducive to identification. With DNA-based techniques gradually perfected and widely used, the integrative (polyphasic) taxonomy approach for species delimitation is recommended, including the combination of genealogy and multiparametric phenotypes [34,35], especially for examining the presence of species complexes and cryptic species [31]. Therefore, we hypothesized that T. harzianum, which was originally identified by ITS sequence and morphology in previous studies, probably belonged to the T. harzianum complex. However, the present study showed that the complex still contained many taxa, indicating that the previous identification was not accurate. Furthermore, it is also difficult to identify species of the Harzianum clade according to exclusive tef1-α or rpb2 sequence data [24,25]. Therefore, the combination of tef1-α and rpb2 sequences for phylogenetic analysis is highly recommended to identify species in the Harzianum clade.
Taxonomy of Trichoderma dates back to the late 18th century [36], and some of them cause economic losses in commercial mushroom farms [37]. Over more than a century, successive findings have brought the number of known species of the genus to over 441 [1,23,38]. Trichoderma species are located throughout the world, and more than 30 of them are mushroom inhabiting (Figure 1, Table 2). They are isolated from the substrate or fruiting bodies of Agaricus bisporus, Lentinula edodes, Pleurotus ostreatus, Ganoderma lingzhi, etc. and are mainly located in the Harzianum, Longibrachiatum, and Viride clades [39]. There may still be many unknown Trichoderma species associated with the growth of edible fungi and their related living environment. The phylogenetical difference between Trichoderma spp. on edible fungi substrates and from other sources deserves further analysis.
Table 2.
Trichoderma spp. associated with the contaminated substrates of edible fungi.
| Species | Cultivated Mushroom | Reference |
|---|---|---|
| T. aggressivum | Agaricus bisporus | [40,41] |
| T. asperellum | A. bisporus | [9,42] |
| T. atroviride | L. edodes, Pleurotus ostreatus, A. bisporus, Ganoderma lingzhi | [8,9,43] |
| T. aureoviride | Auricularia heimuer, Flammulina filiformis, L. edodes | [44] |
| T. breve | L. edodes | [45] |
| T. capillare | Agaricus sp. | [46] |
| T. citrinviride | L. edodes, P. ostreatus | [43,47] |
| T. deliquescens | L. edodes | [11] |
| T. ganodermatigerum | G. sichuanense | [27] |
| T. ghanense | A. bisporus | [9] |
| T. guizhouense | P. ostreatus | [48] |
| T. hamatum | A. bisporus | [49] |
| T. harzianum | L. edodes, A. bisporus, P. ostreatus, Agrocybe aegerita | [43,50] |
| T. hengshanicum | G. lingzhi | [51] |
| T. hirsutum | L. edodes | [45] |
| T. koningii | P. ostreatus, A. bisporus | [37,40] |
| T. koningiopsis | Dictyophora rubrovolvata, P. eryngii | [52,53] |
| T. lentinulae | L. edodes | [24] |
| T. longibrachiatum | L. edodes, P. ostreatus, A. aegerita | [9,43,50] |
| T. oblongisporum | L. edodes | [54] |
| T. patella | P. ostreatus | [55] |
| T. pleuroti | P. ostreatus | [56] |
| T. pleuroticola | P. ostreatus, L. edodes, G. lingzhi | [50,54,56] |
| T. polysporum | L. edodes | [57] |
| T. pseudogelatinosum | L. edodes | [58] |
| T. pseudokoningii | P. ostreatus | [37] |
| T. pseudolacteum | L. edodes | [59] |
| T. pseudostramineum | L. edodes | [58] |
| T. reesei | P. ostreatus | [60] |
| T. stramineum | L. edodes | [57] |
| T. stromaticum | A. bisporus | [49] |
| T. virens | P. ostreatus, A. bisporus | [37,40] |
| T. viride | L. edodes | [54] |
Analysis of the biological characteristics of Trichoderma species from contaminated substrates showed that the optimum growth temperature of many Trichoderma species was generally around 30 °C, which was consistent with the phenomenon that contamination of Trichoderma on edible fungi is more likely to occur at high temperatures. Therefore, reasonable control the growth environment temperature of edible fungi may be a reasonable approach to prevent or delay the outbreak of Trichoderma contamination during production. More broadly, research is also needed to analyze the mechanism of occurrence of Trichoderma spp. contamination, such as the correlation between contamination occurrence and the growth environment of edible fungi.
With the increased number of species joining the Harzianum clade, understanding of Trichoderma spp. will become more sophisticated and intelligible, and reasonable species concepts will be firmly established. Accumulated knowledge of Trichoderma, especially the Harzianum clade, will provide useful information for sufficient utilization of resources and for the prevention of contamination of edible fungi.
5. Conclusions
In this study, 39 strains belonging to 10 Trichoderma species isolated from four kinds of edible fungi in North China were obtained, and three novel species belonging to the Harzianum clade were isolated from the contaminated substrates of Auricularia heimuer and Pholiota adipose. More than 30 mushroom-inhabiting Trichoderma species throughout the world mainly located in the Harzianum, Longibrachiatum, and Viride clades were indicated. This study enrich the biodiversity of Trichoderma and provide important support for systematic development of the Harzianum clade.
Acknowledgments
The authors are thankful to Xing-Hong Li and Wei Zhang for technical assistance and thankful to all the sample collectors in this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8111154/s1. Table S1: Strain information and their accession numbers. Table S2: Comparison of the morphological characteristics of Trichoderma auriculariae and its relatives. Table S3: Comparison of the morphological characteristics of Trichoderma miyunense and its relatives. Table S4: Comparison of the morphological characteristics of Trichoderma pholiotae and its relatives. Table S5: The growth rate of three new species in this study incubated at different temperatures and media.
Author Contributions
Conceptualization, W.-T.Q. and S.-Y.Z.; methodology, W.-T.Q.; software, Z.-J.C. and J.Z.; validation, W.-T.Q. and Y.L.; formal analysis, Z.-J.C.; investigation, Z.-J.C. and S.-X.W.; data curation, Z.-J.C. and J.Z.; writing—original draft preparation, Z.-J.C.; writing—review and editing, W.-T.Q. and S.-Y.Z.; visualization, Z.-J.C. and J.Z.; supervision, W.-T.Q. and S.-X.W.; project administration, W.-T.Q. and Y.L.; funding acquisition, W.-T.Q. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was funded by the Beijing Academy of Agriculture and Forestry Sciences, China (KJCX20220415), the National Natural Science Foundation of China (32002106), and the Rural Revitalization Project of Beijing Municipal Bureau of Agriculture (BJXCZX20221229).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cai F., Druzhinina I.S. In honor of John Bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 2021;107:1–69. doi: 10.1007/s13225-020-00464-4. [DOI] [Google Scholar]
- 2.Druzhinina I.S., Seidl-Seiboth V., Herrera-Estrella A., Horwitz B.A., Kenerley C.M., Monte E., Mukherjee P.K., Zeilinger S., Grigoriev I.V., Kubicek C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 2011;9:749–759. doi: 10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- 3.Nuangmek W., Aiduang W., Kumla J., Lumyong S., Suwannarach N. Evaluation of a newly identified endophytic fungus, Trichoderma phayaoense for plant growth promotion and biological control of gummy stem blight and wilt of muskmelon. Front. Microbiol. 2021;12:634772. doi: 10.3389/fmicb.2021.634772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carillo P., Woo S.L., Comite E., El-Nakhel C., Vinale F. Application of Trichoderma harzianum, 6-pentyl-α-pyrone and plant biopolymer formulations modulate plant metabolism and fruit quality of plum tomatoes. Plants. 2020;9:771. doi: 10.3390/plants9060771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei H., Wu M., Fan A., Su H. Recombinant protein production in the filamentous fungus Trichoderma. Chin. J. Chem. Eng. 2021;30:74–81. doi: 10.1016/j.cjche.2020.11.006. [DOI] [Google Scholar]
- 6.Gupta S., Smith P., Boughton B., Twt R., Sha N., Roessner U. Inoculation of barley with Trichoderma harzianum T-22 modifies lipids and metabolites to improve salt tolerance. J. Exp. Bot. 2021;72:7229–7246. doi: 10.1093/jxb/erab335. [DOI] [PubMed] [Google Scholar]
- 7.Samuel G.J., Dodd S.L., Gams W., Castlebury L.A., Petrini O. Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia. 2002;94:146–170. doi: 10.1080/15572536.2003.11833257. [DOI] [PubMed] [Google Scholar]
- 8.Yan Y., Zhang C., Moodley O., Zhang L., Xu J. Green mold caused by Trichoderma atroviride on the lingzhi medicinal mushroom, Ganoderma lingzhi (Agaricomycetes) Int. J. Med. Mushrooms. 2019;21:515–521. doi: 10.1615/IntJMedMushrooms.2019030352. [DOI] [Google Scholar]
- 9.Hatvani L., Antal Z., Manczinger L., Szekeres A., Druzhinina I.S., Kubicek C.P., Nagy A., Nagy E., Vagvolgyi C., Kredics L. Green mold diseases of Agaricus and Pleurotus spp. are caused by related but phylogenetically different Trichoderma species. Phytopathology. 2007;97:532–537. doi: 10.1094/PHYTO-97-4-0532. [DOI] [PubMed] [Google Scholar]
- 10.Komon-Zelazowska M., Bissett J., Zafari D., Hatvani L., Manczinger L., Woo S., Lorito M., Kredics L., Kubicek C.P., Druzhinina I.S. Genetically closely related but phenotypically divergent Trichoderma species cause green mold disease in oyster mushroom farms worldwide. Appl. Environ. Microb. 2007;73:7415–7426. doi: 10.1128/AEM.01059-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.Y., Yun Y.H., Hyun M.W., Kim M.H., Kim S.H. Identification and characterization of Gliocladium viride isolated from mushroom fly infested oak log beds used for shiitake cultivation. Mycobiology. 2010;38:7–12. doi: 10.4489/MYCO.2010.38.1.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng H., Qiao M., Lv Y.F., Du X., Zhang K.Q., Yu Z.F. New species of Trichoderma isolated as endophytes and saprobes from southwest China. J. Fungi. 2021;7:467. doi: 10.3390/jof7060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ignazio C., Linda M.K. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- 14.Jaklitsch W.M., Komon M., Kubicek C.P., Druzhinina I.S. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia. 2005;97:1365–1378. doi: 10.1080/15572536.2006.11832743. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Z.X., Zhuang W.Y. Trichoderma (Hypocrea) species with green ascospores from China. Persoonia. 2015;34:113–129. doi: 10.3767/003158515X686732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou K., Lu Z., Wu Q., Ni M., Yu C., Wang M., Li Y., Wang X., Xie H., Chen J., et al. MIST: A multilocus identification system for Trichoderma. Appl. Environ. Microb. 2020;86:e01532-20. doi: 10.1128/AEM.01532-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- 19.Higgins D.G., Jeanmougin F., Gibson T.J., Plewniak F., Thompson J.D. The Clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Symp. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swofford D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sinauer Associates; Sunderland, MA, USA: 2002. Version 4.0. [Google Scholar]
- 21.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 22.Nylander J.A.A. MrModeltest v2. Program Distributed by the Author. 2004. [(accessed on 1 January 2018)]. Available online: http://paup.csit.fsu.edu.
- 23.Rambaut A. FigTree. Tree Figure Drawing Tool, v. 1.4.3. 2016. [(accessed on 1 January 2018)]. Available online: http://tree.bio.ed.ac.uk/
- 24.Gu X., Wang R., Sun Q., Wu B., Sun J.-Z. Four new species of Trichoderma in the Harzianum clade from northern China. Mycokeys. 2020;73:109–132. doi: 10.3897/mycokeys.73.51424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaverri P., Branco-Rocha F., Jaklitsch W., Gazis R., Degenkolb T., Samuels G.J. Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia. 2015;107:558–590. doi: 10.3852/14-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan H.S., Lu X., Dai Y.C., Hyde K.V.D., Kan Y.H., Kusan I., He S.H., Liu N.G., Sarma V.V., Zhao C.L., et al. Fungal diversity notes 1277–1386: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2020;104:1–266. doi: 10.1007/s13225-020-00461-7. [DOI] [Google Scholar]
- 27.An X.Y., Cheng G.H., Gao H.X., Li D., Li Y. Phylogenetic analysis of trichoderma species associated with green mold disease on mushrooms and two new pathogens on Ganoderma sichuanense. J. Fungi. 2022;8:704. doi: 10.3390/jof8070704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez M.D.H., Evans H.C., De Abreu L.M., De Macedo D.M., Ndacnou M.K., Bekele K.B., Barreto R.W. New species and records of Trichoderma isolated as mycoparasites and endophytes from cultivated and wild coffee in Africa. Sci. Rep. 2021;11:5671. doi: 10.1038/s41598-021-84111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen K., Zhuang W.Y. Seven new species of Trichoderma from soil in China. Mycosystema. 2017;36:1441–1462. [Google Scholar]
- 30.Chaverri P., Gazis R.O., Samuels G.J. Trichoderma amazonicum, a new endophytic species on Hevea brasiliensis and H. guianensis from the Amazon basin. Mycologia. 2011;103:139–151. doi: 10.3852/10-078. [DOI] [PubMed] [Google Scholar]
- 31.Li Q.R., Tan P., Jiang Y.L., Hyde K.D., Mckenzie E.H.C., Bahkali A.H., Kang J.C., Wang Y. A novel Trichoderma species isolated from soil in Guizhou, T. guizhouense. Mycol. Prog. 2013;12:167–172. doi: 10.1007/s11557-012-0821-2. [DOI] [Google Scholar]
- 32.Chaverri P., Samuels G.J. Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): Species with green ascospores. Stud. Mycol. 2003;48:1–116. [Google Scholar]
- 33.Jaklitsch W.M., Samuels G.J., Dodd S.L., Lu B.S., Druzhinina I.S. Hypocrea rufa/Trichoderma viride: A reassessment, and description of five closely related species with and without warted conidia. Stud. Mycol. 2006;56:135–177. doi: 10.3114/sim.2006.56.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luecking R., Aime M.C., Robbertse B., Miller A.N., Ariyawansa H.A., Aoki T., Cardinali G., Crous P.W., Druzhinina I.S., Geiser D.M., et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 2020;11:14. doi: 10.1186/s43008-020-00033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Druzhinina I., Kubicek C.P. Species concepts and biodiversity in Trichoderma and Hypocrea: From aggregate species to species clusters? J. Zhejiang Univ. Sci. B. 2005;6:100–112. doi: 10.1631/jzus.2005.B0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persoon C.H. Neurospora genetic nomenclature. Romers Neues Mag. Bot. 1794;1:81–128. [Google Scholar]
- 37.Jhune C.S., Leem H.T., Park H.S., Lee C.J., Weon H.Y., Seok S.J., Yoo K.H., Sung G.H. Identification of oyster mushroom green mold pathogen that causes and pathological characteristics. J. Mushrooms. 2014;12:132–137. doi: 10.14480/JM.2014.12.2.132. [DOI] [Google Scholar]
- 38.Barrera V.A., Iannone L., Romero A.I., Chaverri P. Expanding the Trichoderma harzianum species complex: Three new species from Argentine natural and cultivated ecosystems. Mycologia. 2021;113:1136–1155. doi: 10.1080/00275514.2021.1947641. [DOI] [PubMed] [Google Scholar]
- 39.Allaga H., Zhumakayev A., Buchner R., Kocsube S., Szucs A., Vagvolgyi C., Kredics L., Hatvani L. Members of the Trichoderma harzianum species complex with mushroom pathogenic potential. Agronomy. 2021;11:2434. doi: 10.3390/agronomy11122434. [DOI] [Google Scholar]
- 40.Kosanovic D., Potocnik I., Duduk B., Vukojevic J., Stajic M., Rekanovic E., Milijasevic-Marcic S. Trichoderma species on Agaricus bisporus farms in Serbia and their biocontrol. Ann. Appl. Biol. 2013;163:218–230. doi: 10.1111/aab.12048. [DOI] [Google Scholar]
- 41.Marik T., Urban P., Tyagi C., Szekeres A., Leitgeb B., Vagvolgyi M., Manczinger L., Druzhinina I.S., Vagvolgyi C., Kredics L. Diversity profile and dynamics of peptaibols produced by green mould Trichoderma species in interactions with their hosts Agaricus bisporus and Pleurotus ostreatus. Chem. Biodivers. 2017;14:e1700033. doi: 10.1002/cbdv.201700033. [DOI] [PubMed] [Google Scholar]
- 42.Wu X.J., Hu F.P., He H.Z., Xie B.G. Identification of Trichoderma species associated with cultivated edible fungi. J. Agric. Biotechnol. 2008;16:1048–1055. [Google Scholar]
- 43.Kim C.S., Park M.S., Kim S.C., Maekawa N., Yu S.H. Identification of Trichoderma, a competitor of shiitake mushroom (Lentinula edodes), and competition between Lentinula edodes and Trichoderma species in Korea. Plant Pathol. J. 2012;28:137–148. doi: 10.5423/PPJ.2012.28.2.137. [DOI] [Google Scholar]
- 44.Cui L.H. Master’s Thesis. Liaoning Normal University; Dalian, China: 2017. Isolation, Identification and Diversity Analysis of the Contaminating Fungi from the Edible Mushroom-Growing Synthetic Wood Logs. [Google Scholar]
- 45.Wang Y. Master’s Thesis. Guizhou University; Guiyang, China: 2021. Identification of the Pathogen of Lentinus edodes Sticks Rot and Preliminary Study on Its Occurrence. [Google Scholar]
- 46.Samuels G.J., Ismaiel A., Mulaw T.B., Szakacs G., Druzhinina I.S., Kubicek C.P., Jaklitsch W.M. The Longibrachiatum clade of Trichoderma: A revision with new species. Fungal Divers. 2012;55:77–108. doi: 10.1007/s13225-012-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park M.S., Seo G.S., Bae K.S., Yu S.H. Characterization of Trichoderma spp. associated with green mold of oyster mushroom by PCR-RFLP and sequence analysis of ITS regions of rDNA. Plant Pathol. J. 2005;21:229–236. doi: 10.5423/PPJ.2005.21.3.229. [DOI] [Google Scholar]
- 48.Innocenti G., Montanari M., Righini H., Roberti R. Trichoderma species associated with green mould disease of Pleurotus ostreatus and their sensitivity to prochloraz. Plant Pathol. 2019;68:392–398. doi: 10.1111/ppa.12953. [DOI] [Google Scholar]
- 49.Song X.X., Wang Q., Jun J.X., Zhang J.J., Chen H., Chen M.J., Huang J.C., Xie B.Q. Study on accurate identification of four Trichoderma diseases in Agaricus bisporus in factory cultivation. Edible Fungi. 2019;41:67–72. [Google Scholar]
- 50.Choi I.Y., Choi J.N., Hyu L.W., Sharma P.K. Isolation and identification of mushroom pathogens from Agrocybe aegerita. Mycobiology. 2010;38:310–315. doi: 10.4489/MYCO.2010.38.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai M., Idrees M., Zhou Y., Zhang C., Xu J. First report of green mold disease caused by Trichoderma hengshanicum on Ganoderma lingzhi. Mycobiology. 2020;48:427–430. doi: 10.1080/12298093.2020.1794230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., Zhou X., Zhao J., Tang X., Pasquali M., Migheli Q., Berg G., Cernava T. Occurrence of green mold disease on Dictyophora rubrovolvata caused by Trichoderma koningiopsis. J. Plant Pathol. 2021;103:981–984. doi: 10.1007/s42161-021-00861-x. [DOI] [Google Scholar]
- 53.Kim S.W., Kim S., Lee H.J., Park J.W., Ro H.S. Isolation of fungal pathogens to an edible mushroom, Pleurotus eryngii, and development of specific ITS primers. Mycobiology. 2013;41:252–255. doi: 10.5941/MYCO.2013.41.4.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G., Cao X., Ma X., Guo M., Liu C., Yan L., Bian Y. Diversity and effect of Trichoderma spp. associated with green mold disease on Lentinula edodes in China. Microbiologyopen. 2016;5:709–718. doi: 10.1002/mbo3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hua R., Li J.Y., Liu S.X., Luo X.K., Wang X.Y., Liu C.L., Zhang L.Y., Sun D.F. Investigation on common dieases of Pleurotus ostreatus and identification of pathogens. Edible Fungi China. 2021;40:80–86. [Google Scholar]
- 56.Park M.S., Bae K.S., Yu S.H. Two new species of Trichoderma associated with green mold of oyster mushroom cultivation in Korea. Mycobiology. 2006;34:111–113. doi: 10.4489/MYCO.2006.34.3.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyazaki K., Tsuchiya Y., Okuda T. Specific PCR assays for the detection of Trichoderma harzianum causing green mold disease during mushroom cultivation. Mycoscience. 2009;50:94–99. doi: 10.1007/S10267-008-0460-2. [DOI] [Google Scholar]
- 58.Kim C.S., Yu S.H., Nakagiri A., Shirouzu T., Sotome K., Kim S.C., Maekawa N. Re-evaluation of Hypocrea pseudogelatinosa and H. pseudostraminea isolated from shiitake mushroom (Lentinula edodes) cultivation in Korea and Japan. Plant Pathol. J. 2012;28:341–356. doi: 10.5423/PPJ.OA.05.2012.0068. [DOI] [Google Scholar]
- 59.Kim C.S., Shirouzu T., Nakagiri A., Sotome K., Maekawa N. Trichoderma eijii and T. pseudolacteum, two new species from Japan. Mycol. Prog. 2013;12:739–753. doi: 10.1007/s11557-012-0886-y. [DOI] [Google Scholar]
- 60.Lan B.M. Isolation and identification of Trichoderma on oyster mushroom in Quanzhou. Chin. J. Trop. Agric. 2022;42:81–85. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.