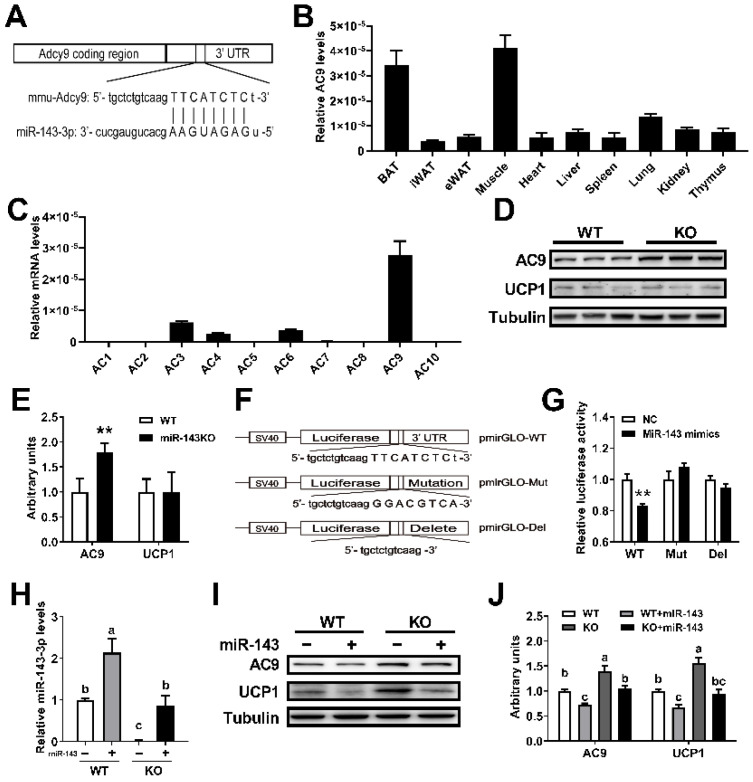
Figure 6.
miR-143 targets AC9. (A) The predicted target site for miR-143 in the 3′-UTR of AC9. (B) The tissue distribution of mRNA encoding AC9 (n = 6–8). (C) The type 1 to 10 adenylyl cyclase mRNA expression levels in BAT (n = 6–8). (D,E) The AC9 and UCP1 protein expression levels in the BAT of WT and KO mice fed ND (n = 3). (F) AC9 3′-UTR with fully changed nucleotides in the seed sequence or with a deleted seed sequence disrupts the binding of miR-143. (G) Three 3′-UTR sequences were respectively cloned into the pGL3-Control vector downstream of the luc+ gene, and luciferase assays with the constructed vectors were transfected into differentiated 3T3-L1 cells with negative control (NC) or miR-143 mimic were performed (n = 8–10). (H–J) The miR-143 (H) (n = 4) and AC9 protein (I,J) (n = 3) expression levels in WT or KO brown adipocytes treated with blank or miR-143 expression lentivirus. Data are presented as the mean ± SEM. ** p < 0.01 vs. controls. Different letters above groups represent significant differences, with shared letters representing no significant differences, as determined by a two-tailed unpaired Student’s t-test (E,G) and two-way ANOVA followed by Tukey’s post hoc test (H,J).