Figure 3.
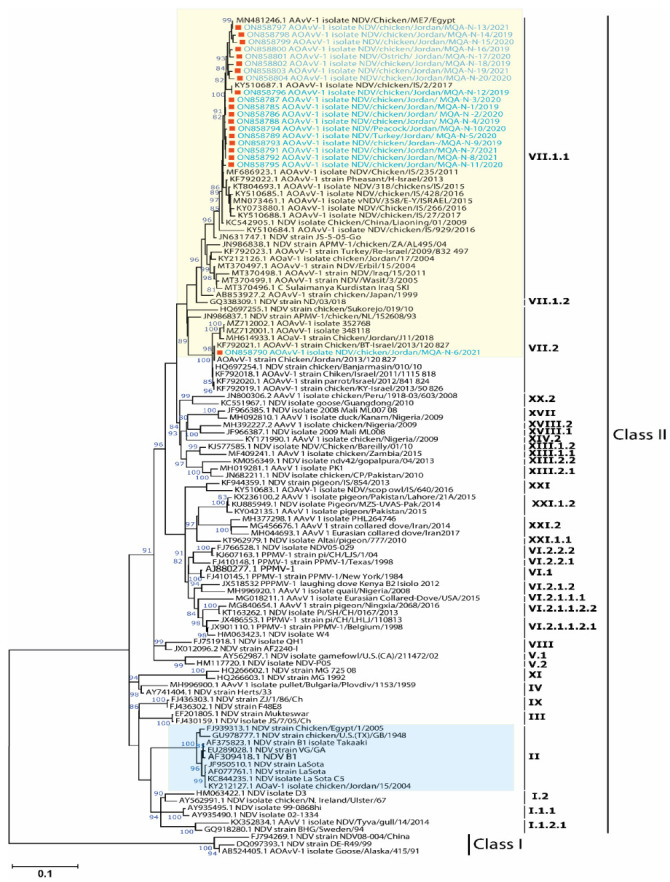
Phylogenetic analysis of the studied AOAvV-1 isolates and their clustering patterns with representative AOAvV-1 isolates. Full-length F-gene (1662 nt)-based phylogenetic analysis of our AOAvV-1 isolates with representative strains of each genotype. Reported isolates clustered in the genotype VII.1.1 of class II. Unrooted phylogenetic trees were generated using the distance-based using maximum likelihood method and MEGA 6 software. Statistical support for tree branches was assessed by bootstrap analysis using 1000 replications of bootstrap re-sampling; numbers above branches indicate neighbor-joining bootstrap values that were ≥80%; the tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The reported AOAvV-1 isolates in this study are marked with red square within yellow box, however, NDV genotype II including LaSota (commonly used vaccine) was labelled within light blue box.
