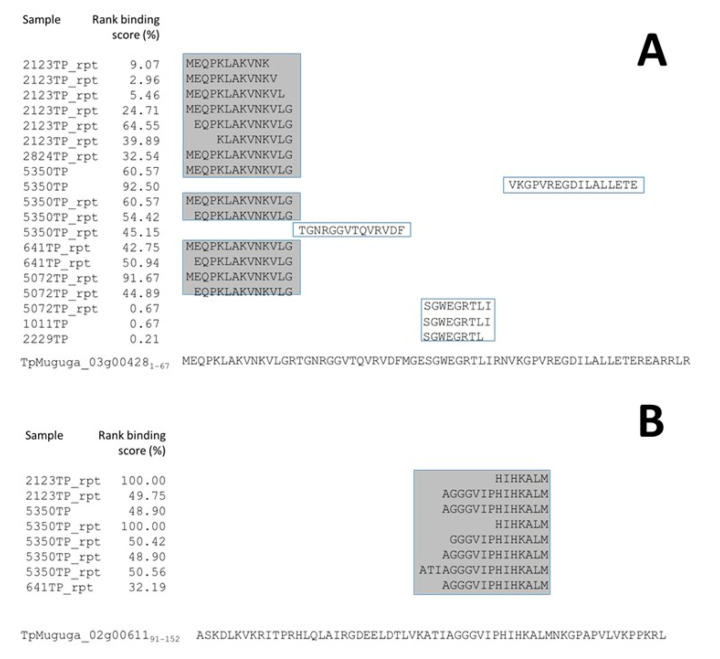
Figure 3.
Alignment of peptides identified from (A) TpMuguga_03g00428 (ribosomal protein S28-B40S) and (B) TpMuguga_02g00611 (histone H2A variant 1) in BoLA-I elution datasets. (A) The 19 peptides identified from 8 different samples are shown aligned against the parent protein. Fourteen of the peptides (from 6 different samples representing 4 different MHCI haplotypes) are found as overlapping peptides in the 1–14 region. The median rank-predicted binding score of these peptides is 43.8% (range = 2.96–64.55%), with none predicted to be BoLA-I binders. The mean length of the peptides was 13 a.a. (B) The 8 peptides identified from 4 different samples (representing 3 different MHCI haplotypes) are shown aligned against the parent protein. All 8 peptides are overlapping peptides in the 121–137 region. The median rank-predicted score of these peptides is 50.1% (range = 32.2–100%), with none predicted to be BoLA-I binders. None of the peptides were of the 8–12 a.a. length of canonical MHCI-binding peptides.