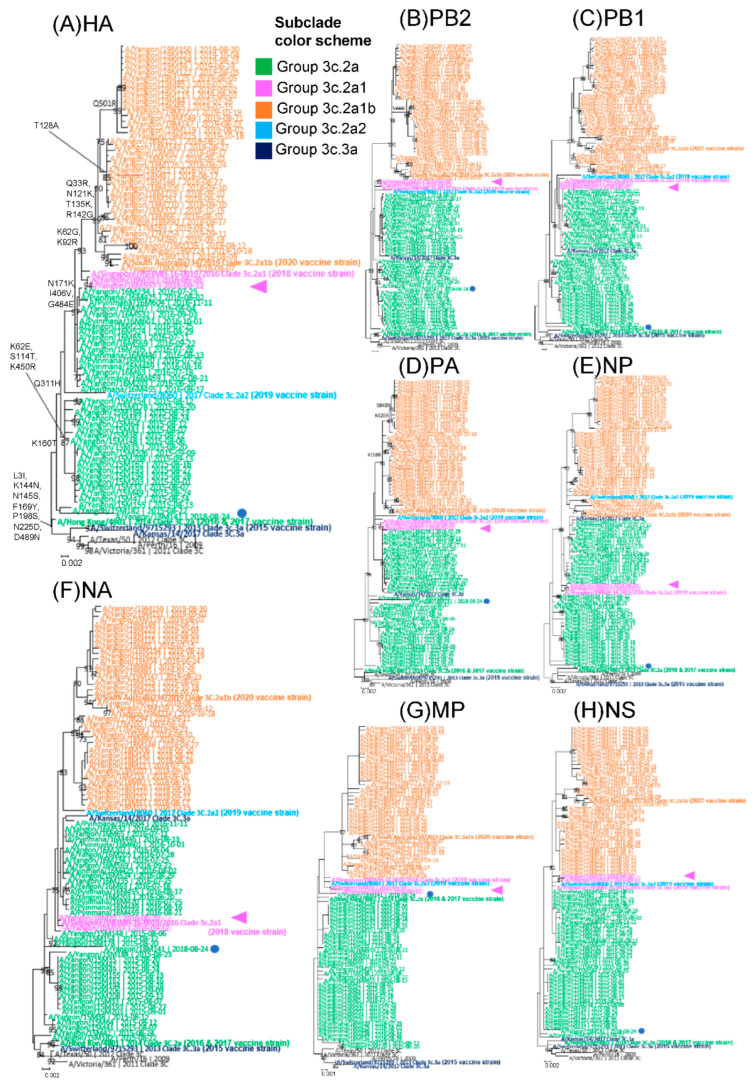
Figure 1.
Maximum-likelihood phylogenetic trees of all eight segments of influenza A/H3N2 viruses circulating in Myanmar and comparison of sequences from 79 strains isolated in Myanmar between 2015 and 2019 and to the Southern Hemisphere vaccine strains of known clades recommended by the WHO. Southern Hemisphere vaccine strains from 2015 to 2019 are indicated by bold letters according to clade classification by the WHO. (A) HA, (B) PB2, (C) PB1, (D) PA, (E) NP, (F) NA, (G) MP, and (H) NS segments. Amino acid substitutions compared to A/Texas/50/2012 were indicated in HA. The two strains that belonged to 3c.2a1 are highlighted in pink and denoted by pink triangles to clarify the topologies concerning 3c.2a and 3c.2a1b viruses. One revival strain, A/Yangon/18M141/2018, is marked with blue circles. The phylogenetic tree was inferred by the maximum-likelihood method using 1000 bootstrap replicates implemented in MEGA v.7.0.26. Branch values > 70% are indicated at the nodes.