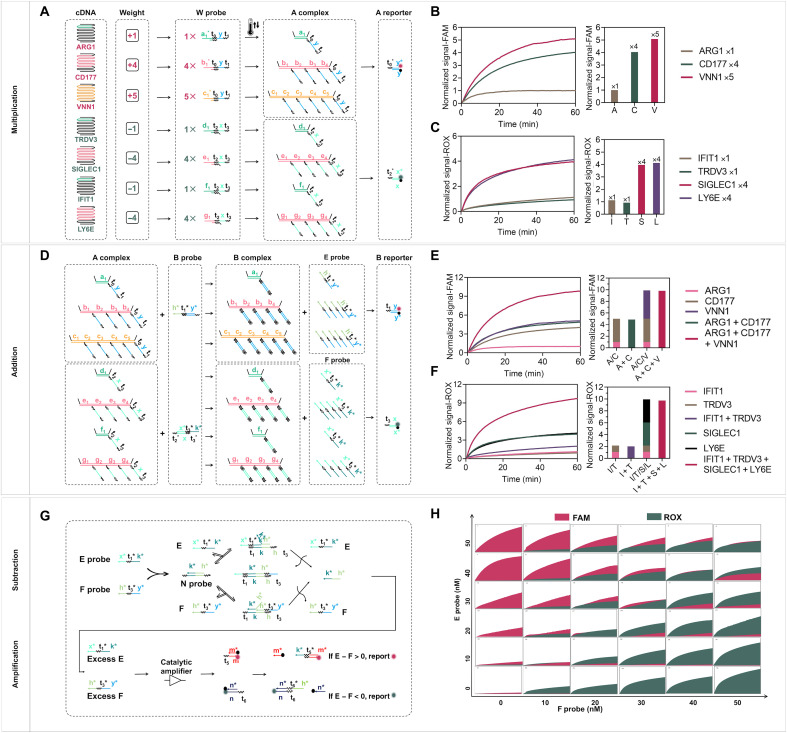
Fig. 3. Validation of the DNA molecular classifier.
(A) Scheme for multiplication [Wn × c(cDNA)n = An]. A reporter is used to verify the multiplication results. (B) Fluorescence kinetics of 10 nM cDNA of ARG1, CD177, and VNN1 genes in the FAM channel (Wn = 1, 4, and 5). (C) Fluorescence kinetics of 10 nM cDNA of IFIT1, TRDV3, SIGLEC1, and LY6E genes with different weights in the ROX channel (Wn = 1, 1, 4, and 4). (D) Scheme for summation (E = A1 + A2 + A3; F = A4 + A5 + A6 + A7). (E) Fluorescence kinetics of A1, A2, and A3 alone and their sums (A1 + A2, A1 + A2 + A3) in the presence of equivalent B probes and B reporter in the FAM channel (i.e., 1, 4, 5, 1 + 4, 1 + 5, and 4 + 5). The normalized FAM fluorescence at 60 min was used to compare the individual and summation signals. (F) Fluorescence kinetics of A4, A5, A6, and A7 alone and their sums (A4 + A5 and A4 + A5 + A6 + A7) in the presence of equivalent B probes and B reporter (i.e., 1, 4, 5, 1 + 4, 1 + 5, and 4 + 5). The normalized ROX fluorescence at 60 min was used to compare the individual and summation signals. (G) Scheme for subtraction (diagnostic result = E − F) and amplification step for sensitivity improvement. Excess E and F probes can be reported by FAM and ROX, respectively. (H) Winner-take-all behavior for different ratios of E and F. The x axis of each small graph is time (from 0 to 60 min), and the y axis is the relative fluorescence units. Samples resulting in a normalized signal of [FAM] − [ROX] > 0 belong to the bacterial infection category, and samples with [FAM] − [ROX] < 0 belong to the viral infection category.