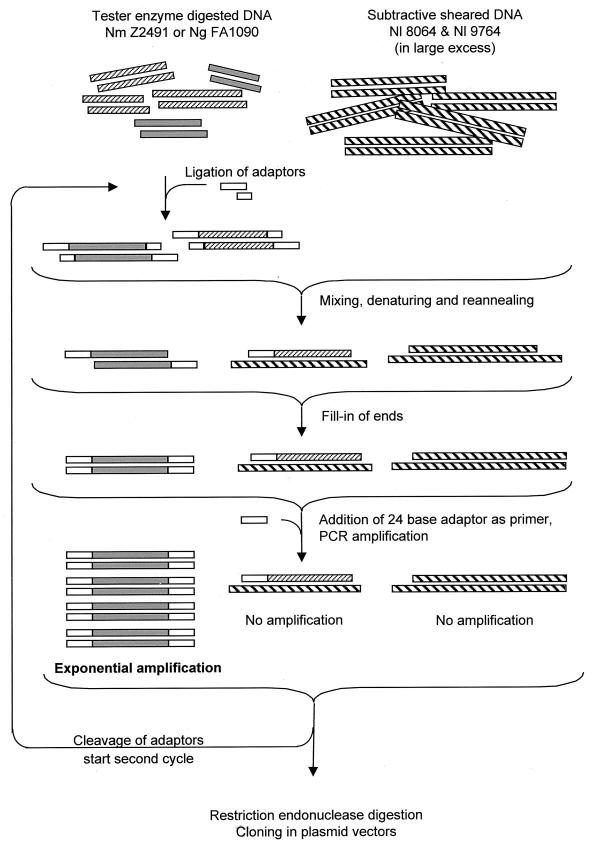
FIG. 1.
Procedure for representational difference analysis. Sequences specific to the pathogen are represented in grey; those in common with N. lactamica are hatched. DNA from N. meningitidis or from N. gonorrhoeae was digested with frequently cutting restriction endonucleases and ligated to adapter pairs such that only the 5′ end of each DNA stand was covalently linked to the 24-bp adapter. On denaturing, mixing, and reannealing, only the (pathogen-specific) sequences with an adapter covalently linked were able to rehybridize with their complementary sequence. The fragments of randomly sheared N. lactamica chromosome are generally over 10 times as long as the restriction fragments from the pathogen and not only sequester all pathogen fragments having homologies in N. lactamica but also, in the large majority of cases, prevent the polymerase from synthesizing the complement of the adapter during the filling-in procedure. Hence, these common fragments are effectively prevented from being amplified, and only the pathogen-specific fragments possessing an adapter at each end can be exponentially amplified.