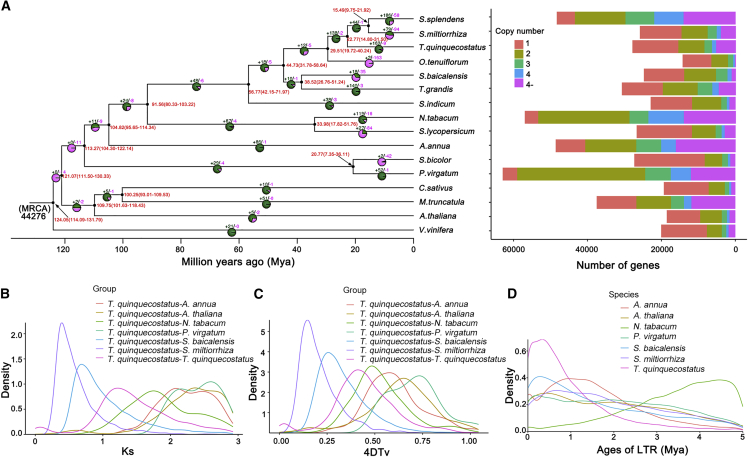
Figure 3.
Genome comparison and evolutionary analysis.
(A) Phylogenetic analysis, divergence time estimates, and the number of gene copies and their distribution among 16 plant species. The tree was constructed on the basis of 951 single-copy truly orthologous genes. Divergence times (Mya) are indicated by the red numbers beside the branch nodes. The numbers of gene-family expansion and contraction events are indicated by green and pink numbers, respectively, on each species branch. MRCA, most recent common ancestor.
(B) Distribution of the synonymous substitution rate (Ks) between T. quinquecostatus and A. annua, A. thaliana, N. tabacum, P. virgatum, S. baicalensis, S. miltiorrhiza, and T. quinquecostatus.
(C) Genome duplication in T. quinquecostatus and genomes of related species as revealed by 4DTv analyses.
(D) Distribution of insertion ages of LTR-retrotransposons in the genomes of T. quinquecostatus and genomes of related species. LTR, long terminal repeat; Mya, million years ago.