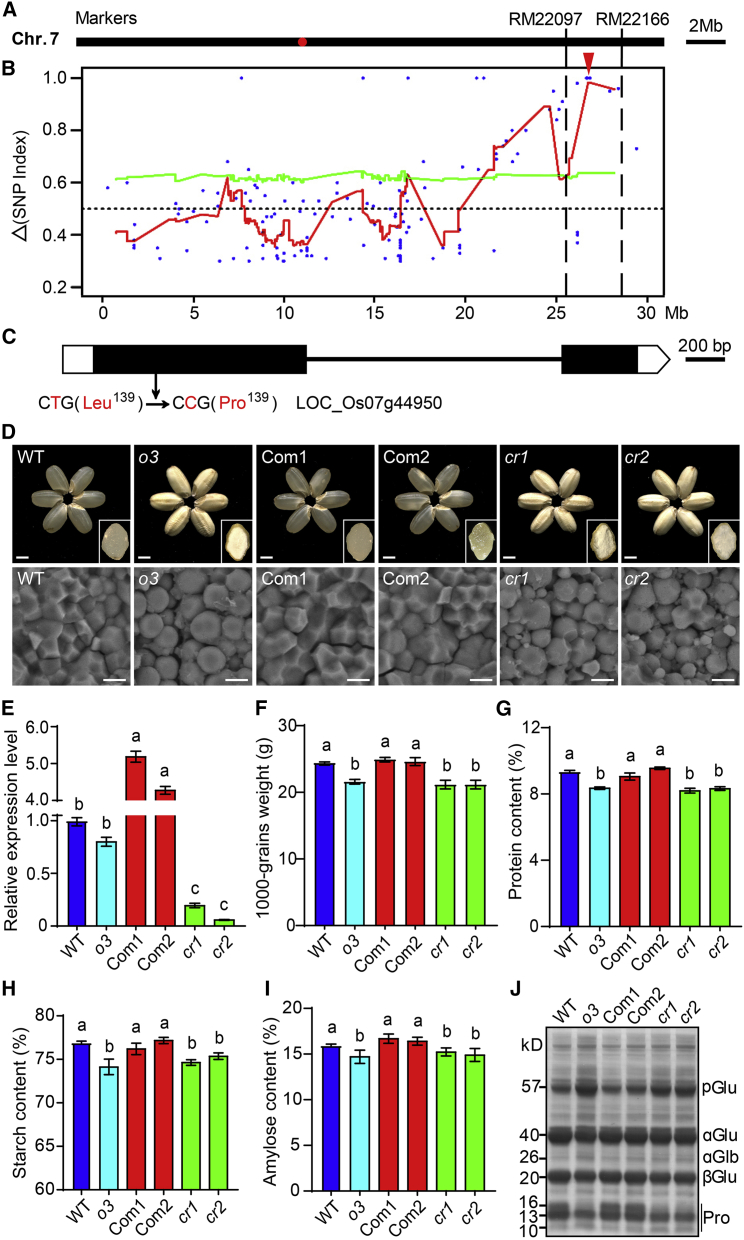
Figure 3.
Identification of Opaque3 (O3) using Map-based cloning and the MutMap method.
(A) The O3 locus was first mapped to the long arm of chromosome 7 between the markers RM22097 and RM22166.
(B) Identification of genomic regions possibly harboring causal mutations for o3 mutants using MutMap. The red curves represent SNP index plots on chromosome 7; the red arrowhead indicates a single peak detected in the primary mapping region.
(C) Gene structure and mutation site in O3 (LOC_Os07g44950). A SNP (T to C) caused a change from Leu-139 to Pro-139 in the encoded protein.
(D) Grain appearance and SG morphology of the WT, o3 mutant, complementation lines (o3 mutant expressing O3; Com1 and Com2), and O3 knockout lines (cr1 and cr2). Insets show transverse sections of representative grains. Scale bars, 2 mm (top) and 5 μm (bottom).
(E) Relative expression level of O3 in developing endosperm of WT, o3, Com1, Com2, cr1, and cr2.
(F–I) 1000-grain weight (F), total protein content (G), total starch content (H), and amylose content (I) of WT, o3, Com1, Com2, cr1, and cr2 grains.
Data in (E–I) are means ± SD from at least three biological replicates. Significant differences are indicated by different letters according to Student’s t-test.
(J) SDS-PAGE profiles of total storage proteins of WT, o3, Com1, Com2, cr1, and cr2 dry grains.