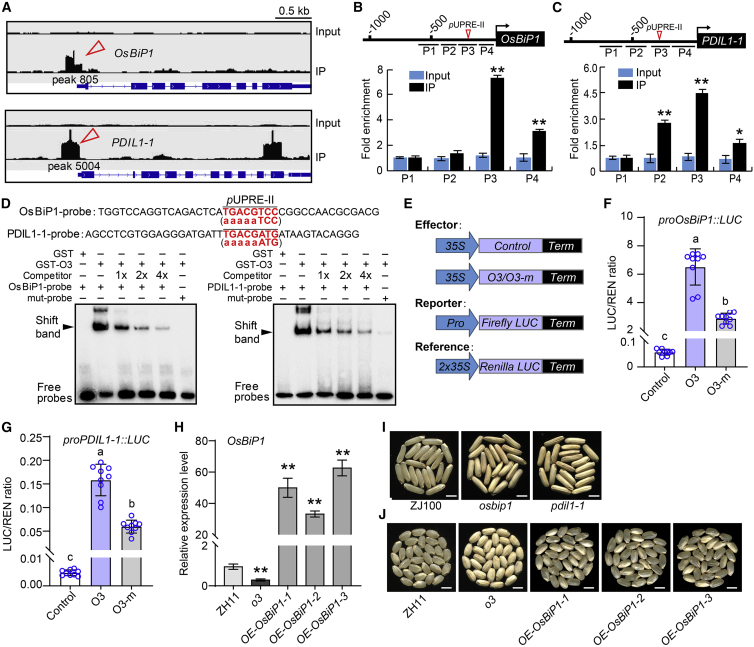
Figure 5.
O3 directly binds to the promoters of OsBIP1 and PDIL1-1 to activate their transcription.
(A) ChIP-seq results showing the distribution of O3 binding sites for OsBIP1 and PDIL1-1 loci, as shown in Integrative Genomics Viewer. Red arrowheads indicate significant peaks calculated by MACS2; positions of peaks are shown in Attachment data II. The input sample was used as a negative control.
(B and C) ChIP-qPCR assay showing the enrichment of O3 at promoter regions of OsBIP1(B) and PDIL1-1(C). DNA samples acquired before immunoprecipitation were used as the input.
(D) EMSA showing that O3 can bind to probes of OsBIP1 and PDIL1-1. The 5-bp consensus pUPRE-II sequence (TGACG) in the promoters of OsBIP1 and PDIL1-1 is indicated by red text. The mutated pUPRE-II motif in mut probes is showed in brackets.
(E–G) LUC transient transactivation assay in rice protoplasts. Constructs used in the transient expression assays are shown in (E). O3 significantly activated transcription of OsBIP1(F) and PDIL1-1(G). O3-m represents the mutant form with a Leu-139 to Pro-139 substitution in the coding region.
(H) qRT–PCR analysis of OsBIP1 transcript level in OE (OE-OsBIP1) lines in the o3 mutant background.
(I) The grain appearance of osbip1 and pdil1-1 mutants in the ZhongJian100 (ZJ100) background. Scale bars, 5 mm.
(J) The grain appearance of OE-OsBIP1 lines in the o3 background. Scale bars, 5 mm.
Data in (B), (C), and (F–H) are means ± SD from at least three biological replicates. Statistically significant differences were determined using Student’s t-test (indicated by different lowercase letters (P < 0.05); ∗P < 0.05, ∗∗P < 0.01).