Figure 3.
Adaptive changes in translation initiation factors and ribosomal proteins to combat protein synthesis inhibition induced by high pressure and low temperature in the hadal zone
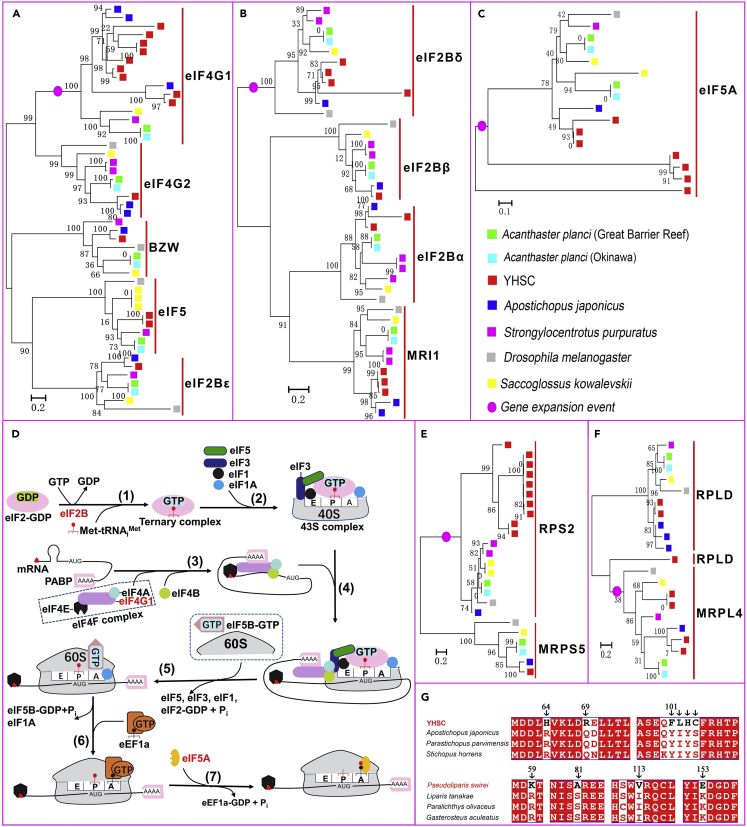
(A–C) Phylogenetic trees showing the expansion of genes for the translation initiation factors eIF4G1, eIF2Bδ, and eIF5A in the YHSC genome.
(D) Model of the translation initiation pathways.57,58,59,60 Expanded genes are marked in red. (1) eIF2B promotes the exchange of GDP-GTP on eIF2, a rate-limiting step of translation initiation. Then, eIF2-GTP forms a ternary complex (eIF2-GTP-Met-tRNAiMet) together with initiator tRNA (Met-tRNAiMet). (2) The ternary complex was recruited to 40S subunits to form 43S complexes with other factors. (3) The eIF4F complex, consisting of eIF4E, eIF4A, and eIF4G1, bound to mRNA and mediated the unwinding of the mRNA cap-proximal region with the help of eIF4B, and mRNA was activated. (4) 43S complex attached to the activated mRNA and scans the 5′UTR of mRNA to the initiation codon - AUG. (5) eIF5B-mediated ribosomal subunits joining and displacement of translation initiation factors. (6) eEF1A carries aminoacyl-tRNA to the A site of the ribosome.61 (7) eIF5A mediated the formation of the first peptide bond.59,62
(E and F) Phylogenetic trees based on protein sequences containing domains of Ribosomal_S5 (E), and Ribosomal_L4 (F) show the expansion of RPS2 and MRPL4 genes in the YHSC genome.
(G) Alignment of the amino acid sequence of MRPS27. MRPS27 is a gene that experienced convergent positive selection between the YHSC and P. swirei. Amino acids unique to the YHSC or P. swirei are indicated by arrows.